C4.5,CART,randomforest的实践
Posted 嘟嘟_猪
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了C4.5,CART,randomforest的实践相关的知识,希望对你有一定的参考价值。
#################################Weka-J48(C4.5)#################################
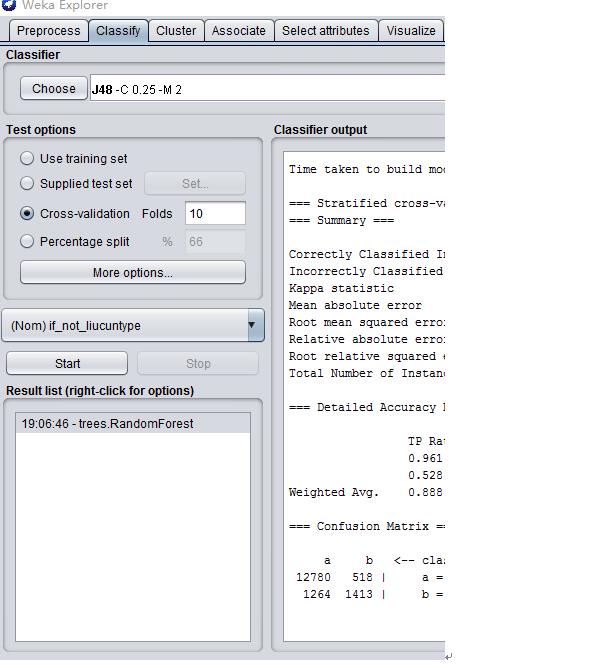
##############################R语言:C4.5######################################
###############################C5.0#############################
data(churn)
treeModel <- C5.0(x = churnTrain[, -20], y = churnTrain$churn)
treeModel
summary(treeModel)
ruleModel <- C5.0(churn ~ ., data = churnTrain, rules = TRUE)
ruleModel
summary(ruleModel)
##################J48#######################
data(iris)
View(iris)
str(iris)
dim(iris)
summary(iris)
#第二步:加载RWeka包
library(RWeka)
#第三步:使用C4.5决策树算法对iris数据集做分类
iris_j48 <- J48(Species ~ ., data = iris)
iris_j48
#第四步:决策树模型摘要分析
summary(iris_j48)
#第五步:模型的可视化
plot(iris_j48)
#第六步:irsi全部数据训练决策树模型的交叉验证
eval_j48 <- evaluate_Weka_classifier(iris_j48, numFolds = 10, complexity = FALSE, seed = 1, class = TRUE)
eval_j48
#第七步:Weka-control的了解
WOW("J48")
#第八步:建立成本敏感决策树分类模型
csc <- CostSensitiveClassifier(Species ~ ., data = iris, control = Weka_control(`cost-matrix` = matrix(c(0, 10, 0, 0, 0, 0, 0, 10, 0), ncol = 3), W = "weka.classifiers.trees.J48", M = TRUE))
eval_csc <- evaluate_Weka_classifier(csc, numFolds = 10, complexity = FALSE, seed = 1, class = TRUE)
eval_csc
##########################R语言:CART##########################################
library(rpart)
sol.rpart<-rpart(Sepal.Length~Sepal.Width+Petal.Length+Petal.Width,data=iris)
plot(sol.rpart,uniform=TRUE,compress=TRUE,lty=3,branch=0.7)
text(sol.rpart,all=TRUE,digits=7,use.n=TRUE,cex=0.9,xpd=TRUE)
####################R语言:RandomForest#################################
library(randomForest)
## Classification:
##data(iris)
set.seed(71)
iris.rf <- randomForest(Species ~ ., data=iris, importance=TRUE,
proximity=TRUE)
print(iris.rf)
## Look at variable importance:
round(importance(iris.rf), 2)
## Do MDS on 1 - proximity:
iris.mds <- cmdscale(1 - iris.rf$proximity, eig=TRUE)
op <- par(pty="s")
pairs(cbind(iris[,1:4], iris.mds$points), cex=0.6, gap=0,
col=c("red", "green", "blue")[as.numeric(iris$Species)],
main="Iris Data: Predictors and MDS of Proximity Based on RandomForest")
par(op)
print(iris.mds$GOF)
## The `unsupervised\' case:
set.seed(17)
iris.urf <- randomForest(iris[, -5])
MDSplot(iris.urf, iris$Species)
## stratified sampling: draw 20, 30, and 20 of the species to grow each tree.
(iris.rf2 <- randomForest(iris[1:4], iris$Species,
sampsize=c(20, 30, 20)))
## Regression:
## data(airquality)
set.seed(131)
ozone.rf <- randomForest(Ozone ~ ., data=airquality, mtry=3,
importance=TRUE, na.action=na.omit)
print(ozone.rf)
## Show "importance" of variables: higher value mean more important:
round(importance(ozone.rf), 2)
## "x" can be a matrix instead of a data frame:
set.seed(17)
x <- matrix(runif(5e2), 100)
y <- gl(2, 50)
(myrf <- randomForest(x, y))
(predict(myrf, x))
## "complicated" formula:
(swiss.rf <- randomForest(sqrt(Fertility) ~ . - Catholic + I(Catholic < 50),
data=swiss))
(predict(swiss.rf, swiss))
## Test use of 32-level factor as a predictor:
set.seed(1)
x <- data.frame(x1=gl(53, 10), x2=runif(530), y=rnorm(530))
(rf1 <- randomForest(x[-3], x[[3]], ntree=10))
## Grow no more than 4 nodes per tree:
(treesize(randomForest(Species ~ ., data=iris, maxnodes=4, ntree=30)))
## test proximity in regression
iris.rrf <- randomForest(iris[-1], iris[[1]], ntree=101, proximity=TRUE, oob.prox=FALSE)
str(iris.rrf$proximity)
########################Weka:RandomForest######################################
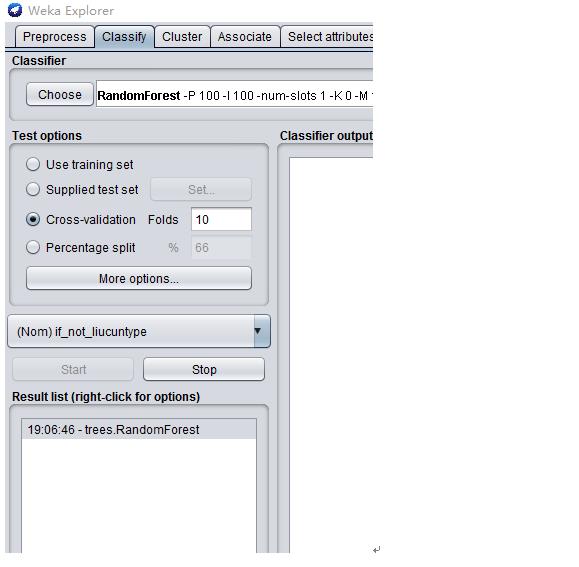
#######################Matlab:RandomForest ###################################
%%%随机森林分类器(Random Forest)
Factor = TreeBagger(nTree, train_data, train_label);
[Predict_label,Scores] = predict(Factor, test_data);
%%%scores是语义向量(概率输出)
############################Python:RandomForest ################################
利用Python的两个模块,分别为pandas和scikit-learn来实现随机森林.
from sklearn.datasets import load_iris
from sklearn.ensemble import RandomForestClassifier
import pandas as pd
import numpy as np
iris = load_iris()
df = pd.DataFrame(iris.data, columns=iris.feature_names)
df[\'is_train\'] = np.random.uniform(0, 1, len(df)) <= .75
df[\'species\'] = pd.Factor(iris.target, iris.target_names)
df.head()
train, test = df[df[\'is_train\']==True], df[df[\'is_train\']==False]
features = df.columns[:4]
clf = RandomForestClassifier(n_jobs=2)
y, _ = pd.factorize(train[\'species\'])
clf.fit(train[features], y)
preds = iris.target_names[clf.predict(test[features])]
pd.crosstab(test[\'species\'], preds, rownames=[\'actual\'], colnames=[\'preds\'])
以上是关于C4.5,CART,randomforest的实践的主要内容,如果未能解决你的问题,请参考以下文章