ANTsPy医学图像配准
Posted junzhaoliang
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了ANTsPy医学图像配准相关的知识,希望对你有一定的参考价值。
ANTsPy主页:https://github.com/ANTsX/ANTsPy
ANTsPy官方文档:https://antspyx.readthedocs.io/_/downloads/en/latest/pdf/
配准ants.registration()
import os
import ants
import numpy as np
import time
fix_path = ‘img_fix.png‘
move_path = ‘img_move.png‘
# 配准所使用的各种方法 各方法耗时:https://www.cnblogs.com/JunzhaoLiang/p/12308200.html
types = [‘Translation‘, ‘Rigid‘, ‘Similarity‘, ‘QuickRigid‘, ‘DenseRigid‘, ‘BOLDRigid‘, ‘Affine‘, ‘AffineFast‘, ‘BOLDAffine‘,
‘TRSAA‘, ‘ElasticSyN‘, ‘SyN‘, ‘SyNRA‘, ‘SyNOnly‘, ‘SyNCC‘, ‘SyNabp‘, ‘SyNBold‘, ‘SyNBoldAff‘, ‘SyNAggro‘, ‘TVMSQ‘]
# 保存为png只支持unsigned char & unsigned short,因此读取时先进行数据类型转换
fix_img = ants.image_read(fix_path, pixeltype=‘unsigned char‘)
move_img = ants.image_read(move_path, pixeltype=‘unsigned char‘)
for t in types:
start = time.time()
out = ants.registration(fix_img, move_img, type_of_transform=t)
reg_img = out[‘warpedmovout‘] # 获取配准结果
reg_img.to_file(t+‘.png‘)
print(t+‘ : ‘, time.time()-start, ‘
‘)
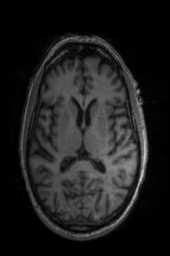
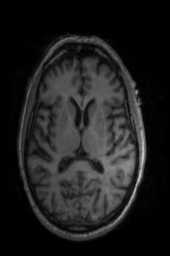
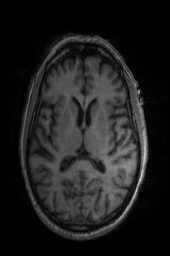
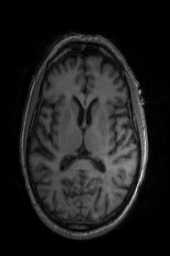
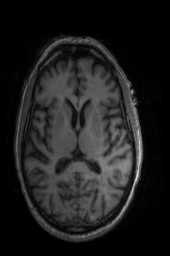
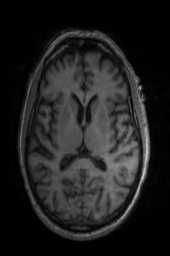
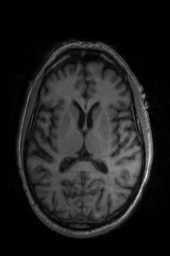
‘Translation‘ ‘Rigid‘ ‘Similarity‘ ‘QuickRigid‘ ‘DenseRigid‘ ‘BOLDRigid‘ ‘Affine‘
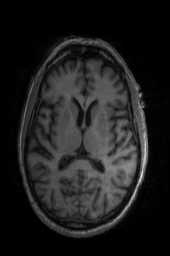
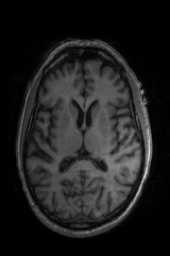
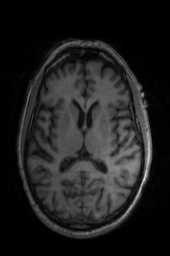
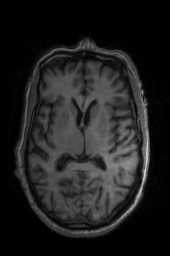
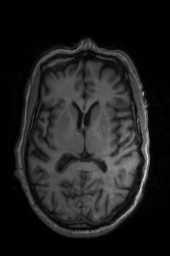
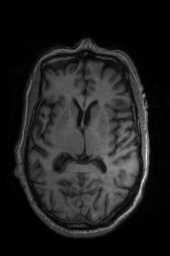
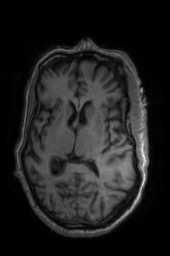
‘AffineFast‘ ‘BOLDAffine‘ #TRSAA# ‘ElasticSyN‘ ‘SyN‘ ‘SyNRA‘ ‘SyNOnly‘
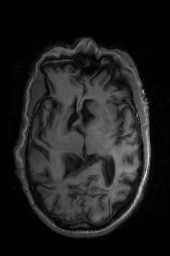
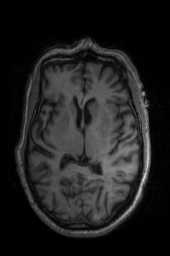
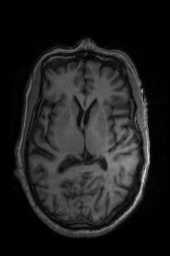
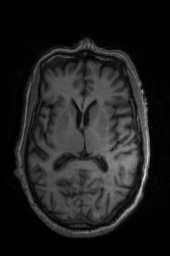
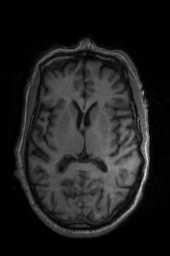
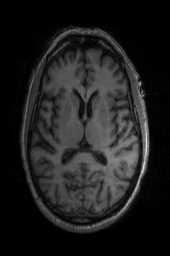
‘SyNCC‘ ‘SyNabp‘ ‘SyNBold‘ ‘SyNBoldAff‘ ‘SyNAggro‘ ‘TVMSQ‘
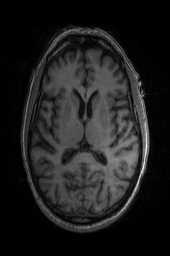
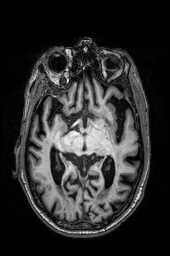
‘img_2‘ ‘img_1‘
校正偏置场ants.n4_bias_field_correction()
import os
import ants
import numpy as npimport time
move_path = ‘img.png‘
move_img = ants.image_read(move_path, pixeltype=‘unsigned char‘)
n4_img = ants.n4_bias_field_correction(move_img)
n4_img = ants.from_numpy(np.array(n4_img.numpy(), dtype=‘uint8‘))
# png只接受 unsigned char & unsigned short
# 保存之前需转换类型
n4_img.to_file(‘TRSAA_n4.png‘)
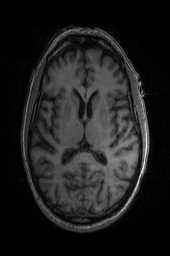
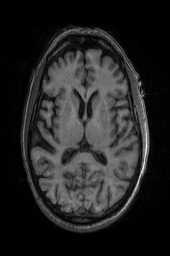
‘OG‘ ‘OG_n4‘ ‘TRSAA‘ ‘TRSAA_n4‘ 以上是关于ANTsPy医学图像配准的主要内容,如果未能解决你的问题,请参考以下文章