威斯康星乳腺癌良性预测
Posted always-fight
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了威斯康星乳腺癌良性预测相关的知识,希望对你有一定的参考价值。
一、获取数据
wget https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data
原始数据以逗号分隔:
各个列的属性:
1.Sample Code Number id number
2.Clump Thickness 1 - 10 肿块厚度
3.Uniformity Of Cell Size 1 - 10 细胞大小均一性
4.Uniformity Of Cell Shape 1 - 10 细胞形状的均一性
5.Marginal Adhesion 1 - 10 边缘附着性
6.Single Epithelial Cell Size 1 - 10 单上皮细胞大小
7.Bare Nuclei 1 - 10 裸核
8.Bland Chromatin 1 - 10 布兰染色质
9.Normal Nucleoli 1 - 10 正常核仁
10.Mitoses 1 - 10 有丝分裂
11.Class 2是良性,4是恶性
二、使用LR和SGD
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import SGDClassifier
from sklearn import metrics
#数据没有标题,因此加上参数header
data = pd.read_csv(‘https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data‘, header=None)
column_names = [‘Sample code number‘,‘Clump Thickness‘,‘Uniformity of Cell Size‘,‘Uniformity of Cell Shape‘, ‘Marginal Adhesion‘,‘Single Epithelial Cell Size‘,‘Bare Nuclei‘, ‘Bland Chromatin‘,‘Normal Nucleoli‘,‘Mitoses‘,‘Class‘]
data.columns = column_names
#发现数据中存在?符号
data = data.replace(to_replace=‘?‘,value = np.nan)
data = data.dropna(how=‘any‘)
#一般1代表恶性,0代表良性(本数据集4恶性,所以将4变成1,将2变成0)
#data[‘Class‘][data[‘Class‘] == 4] = 1
#data[‘Class‘][data[‘Class‘] == 2] = 0
data.loc[data[‘Class‘] == 4, ‘Class‘] = 1
data.loc[data[‘Class‘] == 2, ‘Class‘] = 0
#Sample code number特征对分类没有作用,将数据集75%作为训练集,25%作为测试集
X_train, X_test, y_train, y_test = train_test_split(data[ column_names[1:10] ], data[ column_names[10] ], test_size = 0.25, random_state = 33)
ss = StandardScaler()
X_train = ss.fit_transform(X_train)
X_test = ss.transform(X_test)
lr = LogisticRegression()
lr.fit(X_train, y_train)
lr_y_predict = lr.predict(X_test)
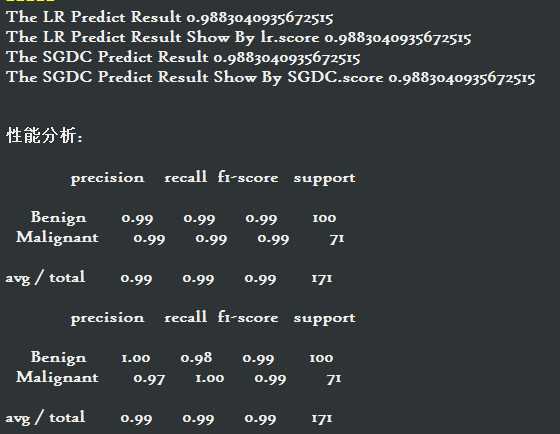
print( ‘The LR Predict Result‘, metrics.accuracy_score(lr_y_predict, y_test) )
#LR也自带了score
print( "The LR Predict Result Show By lr.score", lr.score(X_test, y_test) )
sgdc = SGDClassifier(max_iter = 1000)
sgdc.fit(X_train, y_train)
sgdc_y_predict = sgdc.predict(X_test)
print( "The SGDC Predict Result", metrics.accuracy_score(sgdc_y_predict, y_test) )
#SGDC也自带了score
print( "The SGDC Predict Result Show By SGDC.score", sgdc.score(X_test, y_test) )
print("
")
print("性能分析:
")
#性能分析
from sklearn.metrics import classification_report
#使用classification_report模块获得LR三个指标的结果(召回率,精确率,调和平均数)
print( classification_report( y_test,lr_y_predict,target_names=[‘Benign‘,‘Malignant‘] ) )
##使用classification_report模块获得SGDC三个指标的结果
print( classification_report( y_test,sgdc_y_predict,target_names=[‘Benign‘,‘Malignant‘] ) )
‘‘‘
特点分析:
LR对参数的计算采用精确解析的方法,计算时间长但是模型性能高
SGDC采用随机梯度上升算法估计模型参数,计算时间短但产出的模型性能略低,
一般而言,对于训练数据规模在10万量级以上的数据,考虑到时间的耗用,推荐使用SGDC
‘‘‘
以上是关于威斯康星乳腺癌良性预测的主要内容,如果未能解决你的问题,请参考以下文章
DT:DT实现根据乳腺肿瘤特征向量高精度预测肿瘤的是恶性还是良性
RF:RF实现根据乳腺肿瘤特征向量高精度(better)预测肿瘤的是恶性还是良性—Jason niu