案例:乳腺癌威斯康星州(原始)数据集
Posted 卖山楂啦prss
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了案例:乳腺癌威斯康星州(原始)数据集相关的知识,希望对你有一定的参考价值。
数据:http://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+%28Original%29

import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from pylab import mpl
# 正常显示中文标签
mpl.rcParams['font.sans-serif'] = ['SimHei']
# 正常显示负号
mpl.rcParams['axes.unicode_minus'] = False
# 显示所有列
pd.set_option('display.max_columns', None)
# 显示所有行
pd.set_option('display.max_rows', None)
# 读取数据
df = pd.read_csv(r'C:\\Users\\Administrator\\Desktop\\breast-cancer-wisconsin.csv')
df
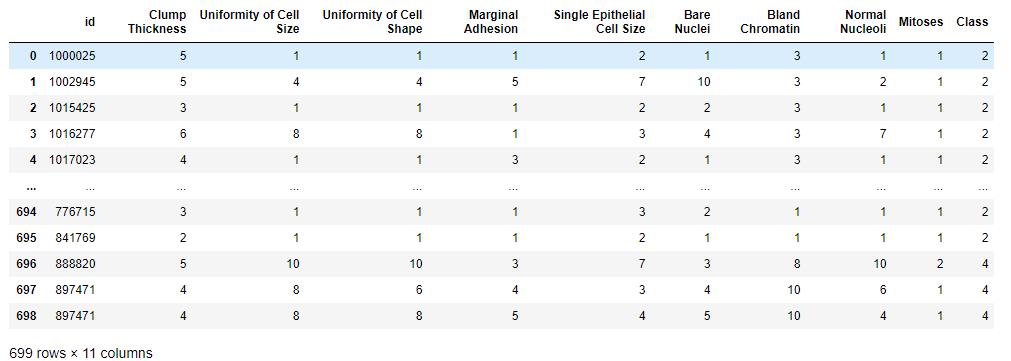
## 通过 .columns 查看列名
df.columns
从df中随机抽取10行
sample1 = df.sample(n=10)
sample1
'''
id Clump Thickness ... Mitoses Class
482 1318169 9 ... 10 4
645 1303489 3 ... 1 2
195 1212422 4 ... 1 2
489 1084139 6 ... 1 4
324 740492 1 ... 1 2
348 832226 3 ... 1 4
112 1172152 10 ... 3 4
124 1175937 5 ... 1 4
273 428903 7 ... 1 4
20 1054590 7 ... 4 4
[10 rows x 11 columns]
'''
df.shape
#去除重复值
df.drop_duplicates(inplace=True)
#恢复索引
df.index = range(df.shape[0])
df.shape#(691, 11)
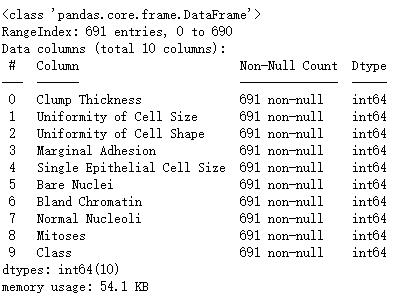
df.info()

object_cols = df.select_dtypes(exclude = 'object').columns
print(object_cols )
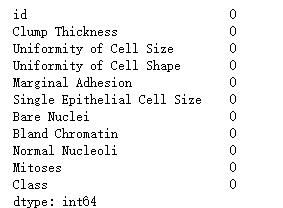
# 缺失情况
df.isnull().sum()
读取数据并做简单的描述性统计 探索性分析,单变量分析,双变量分析 数据与处理和特征工程 建立模型并进行调参,选择最优参数 对测试数据进行预测,并保存模型结果
df.drop(['id'],axis=1,inplace=True)
df
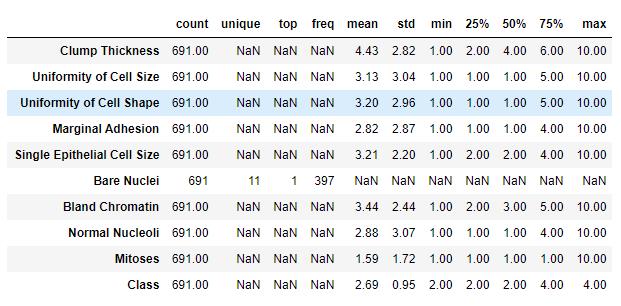
df.describe(include = 'all').T
数据探索性分析
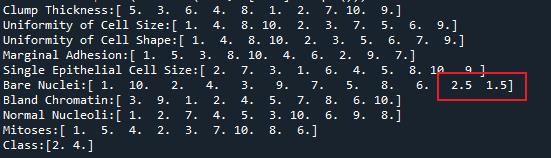
for i, item in enumerate(df):
print('%s:%s'%(item,df[item].unique()))

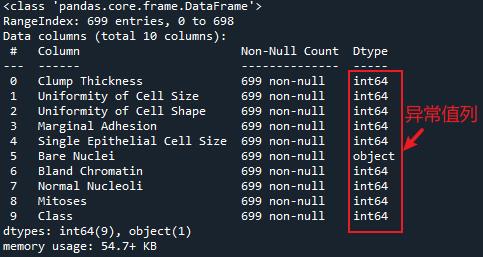
Bare Nuclei列存在一个异常值
df['Bare Nuclei'].value_counts() # 有16个?

# df[df['Bare Nuclei'].isin('?')]
df[df['Bare Nuclei'] =='?']

异常值处理 如果发现了异常值,首先要观察,这个异常值出现的频率 如果异常值只出现了一次,多半是输入错误,直接把异常值删除 如果异常值出现了多次,去跟业务人员沟通,可能这是某种特殊表示,如果是人为造成的错误,异常值留着是没有用 的,只要数据量不是太大,都可以删除 如果异常值占到你总数据量的10%以上了,不能轻易删除。可以考虑把异常值替换成非异常但是非干扰的项,比如说用0 来进行替换,或者把异常当缺失值,用均值或者众数来进行替换
df['Bare Nuclei'].where(df['Bare Nuclei'] != '?',np.nan,inplace=True)
Where用来根据条件替换行或列中的值。如果满足条件,保持原来的值,不满足条件则替换为其他值。默认替换为NaN,也可以指定特殊值。
df['Bare Nuclei'][df['Bare Nuclei'] == '?'] = np.nan
df.head(30)
df.isnull().sum()

拉格朗日插值法—随机森林算法填充–KNN算法—sklearn填充(均值/众数/中位数)
缺失值填补,这里使用众数进行填补
众数
from scipy import stats
stats.mode(df['Bare Nuclei'])[0][0]
'1 ’
df['Bare Nuclei'].fillna(value=stats.mode(df['Bare Nuclei'])[0][0], inplace=True)
df.isnull().sum()

for i, item in enumerate(df):
print('%s:%s'%(item,df[item].unique()))

df['Bare Nuclei'] = df['Bare Nuclei'].astype('int64')
df.info()
题外话:
刚开始我是用KNN插补,结果插补完出现浮点数,这明显不符合数据集的特点(1-10的整数)
对于KNN插补,可以用如下代码
from sklearn.impute import KNNImputer
impute = KNNImputer(n_neighbors = 2)
df_filled = impute.fit_transform(df)# df中有缺失值
df = pd.DataFrame(df_filled,columns=df.columns)
对数据的处理基本完成
数据可视化
sns.set_style("whitegrid") # 使用whitegrid主题
fig,axes=plt.subplots(nrows=5,ncols=2,figsize=(16,20))
for i, item in enumerate(df):
plt.subplot(5,2,(i+1))
#ax=df[item].value_counts().plot(kind = 'bar')
ax=sns.countplot(item,data = df,palette="Pastel1")
plt.xlabel(str(item),fontsize=14)
plt.ylabel('Count',fontsize=14)
plt.xticks(fontsize=13)
plt.yticks(fontsize=13)
#plt.title("Churn by "+ str(item))
i=i+1
plt.tight_layout()
plt.show()
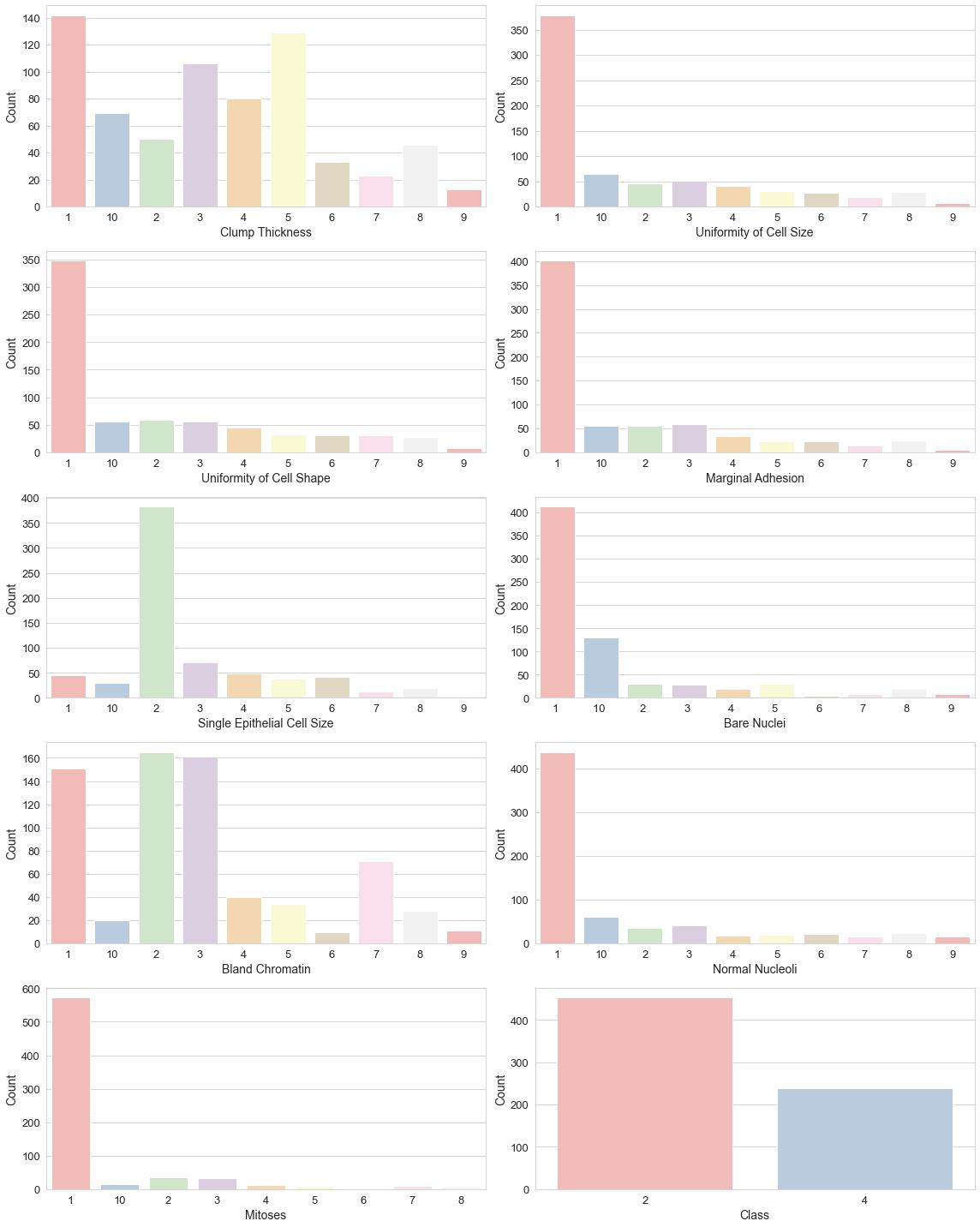
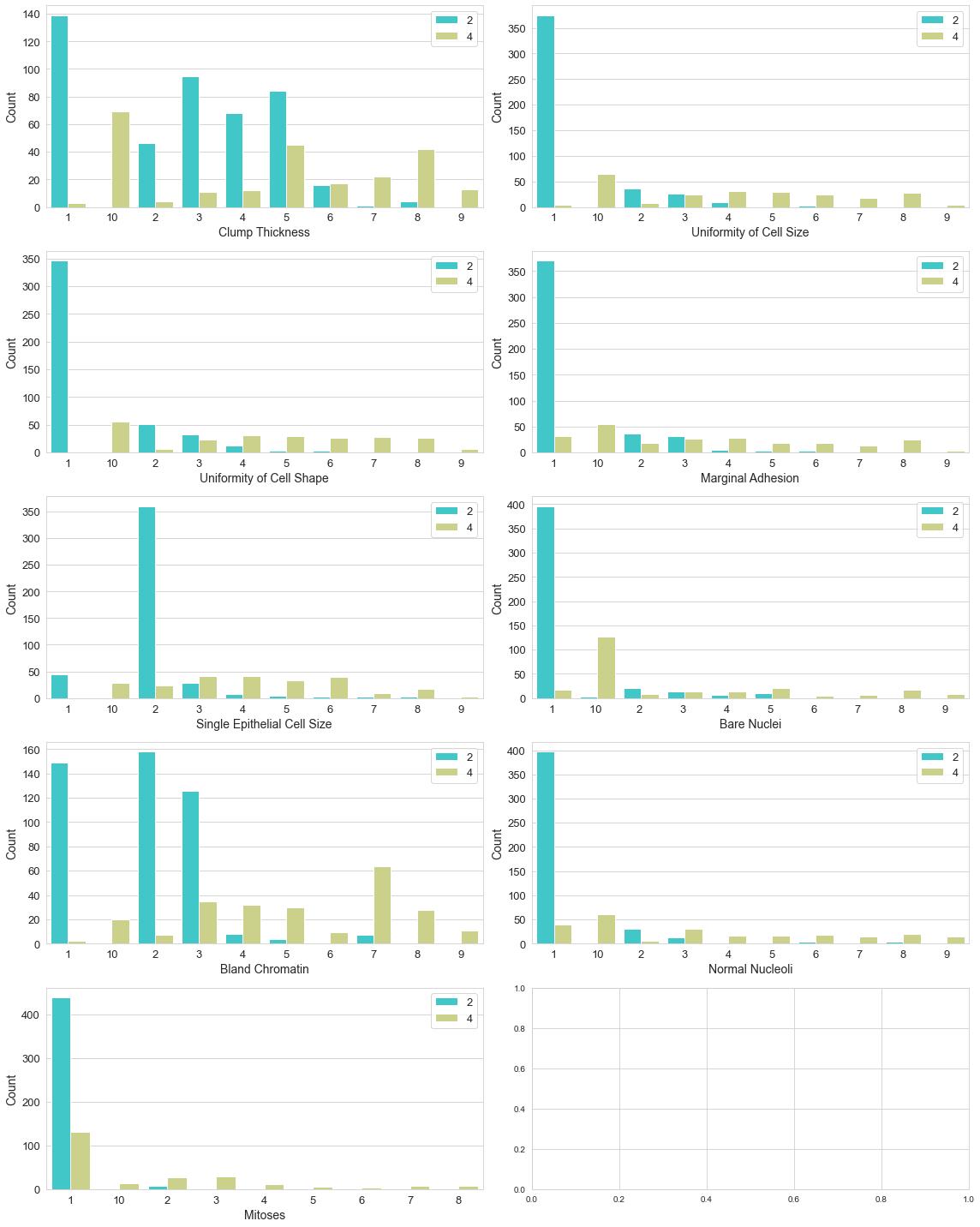
有轻微的样本不均衡问题
fig,axes=plt.subplots(nrows=5,ncols=2,figsize=(16,20))
for i, item in enumerate(df):
if i!=9:
plt.subplot(5,2,(i+1))
ax=sns.countplot(x = item,hue = 'Class',data = df,palette="rainbow")
plt.xlabel(str(item),fontsize=14)
plt.ylabel('Count',fontsize=14)
plt.xticks(fontsize=13)
plt.yticks(fontsize=13)
plt.legend(loc='upper right',fontsize=13)
#plt.title("Churn by "+ str(item))
i=i+1
plt.tight_layout()
plt.show()
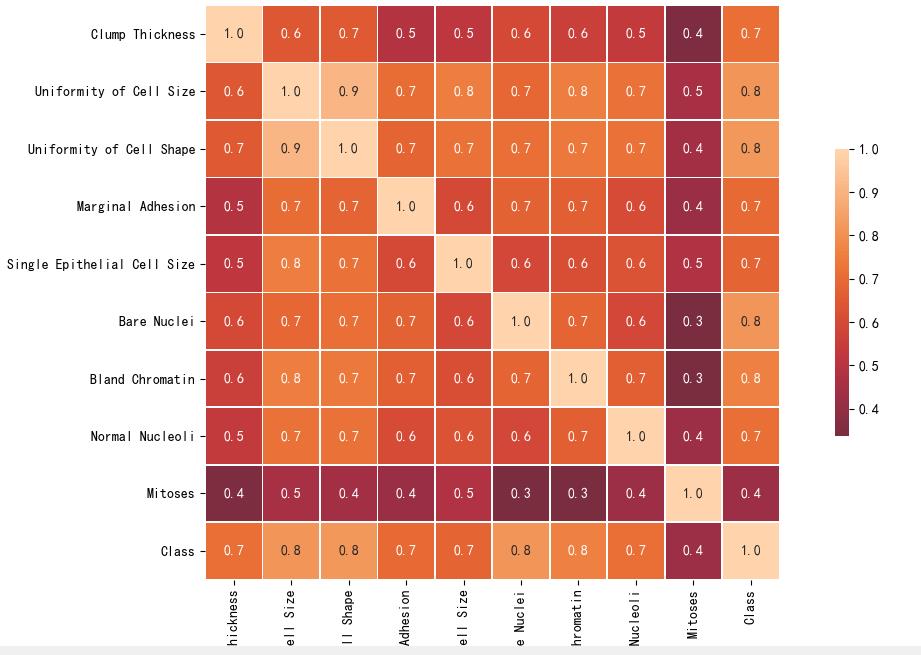
plt.figure(figsize=(12,12))
sns.heatmap(df.corr(), center=0,
square=True, linewidths=.5, cbar_kws="shrink": .5,annot=True, fmt='.1f')
plt.tight_layout()
建模与模型评估
决策树
import pandas as pd
import graphviz
from sklearn import tree
from sklearn.model_selection import train_test_split
#将数据划分为标签和特征
X = df.drop(['Class'],axis = 1)
y = df['Class']
# 分训练集和测试集
Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y,test_size=0.3)
#恢复索引
for i in [Xtrain, Xtest, Ytrain, Ytest]:
i.index = range(i.shape[0])
# 建立模型
clf = tree.DecisionTreeClassifier(criterion="entropy",
random_state=30, # 设置分枝中的随机模式的参数,默认None,在高维度时随机性会表现更明显
splitter="random" # 控制决策树中的随机选项
)
# 训练
clf = clf.fit(Xtrain, Ytrain)
#返回预测的准确度
score = clf.score(Xtest, Ytest)
score # 0.9519230769230769
feature_name = ['团块厚度','细胞大小的均匀性','细胞形状的均匀性','边缘附着力','单层上皮细胞大小','裸核','乏味染色质','正常核仁','线粒体']
dot_data = tree.export_graphviz(clf,out_file = None,
feature_names= feature_name,
class_names=["良性","恶性"],
filled=True,
rounded=True)
graph = graphviz.Source(dot_data)
graph
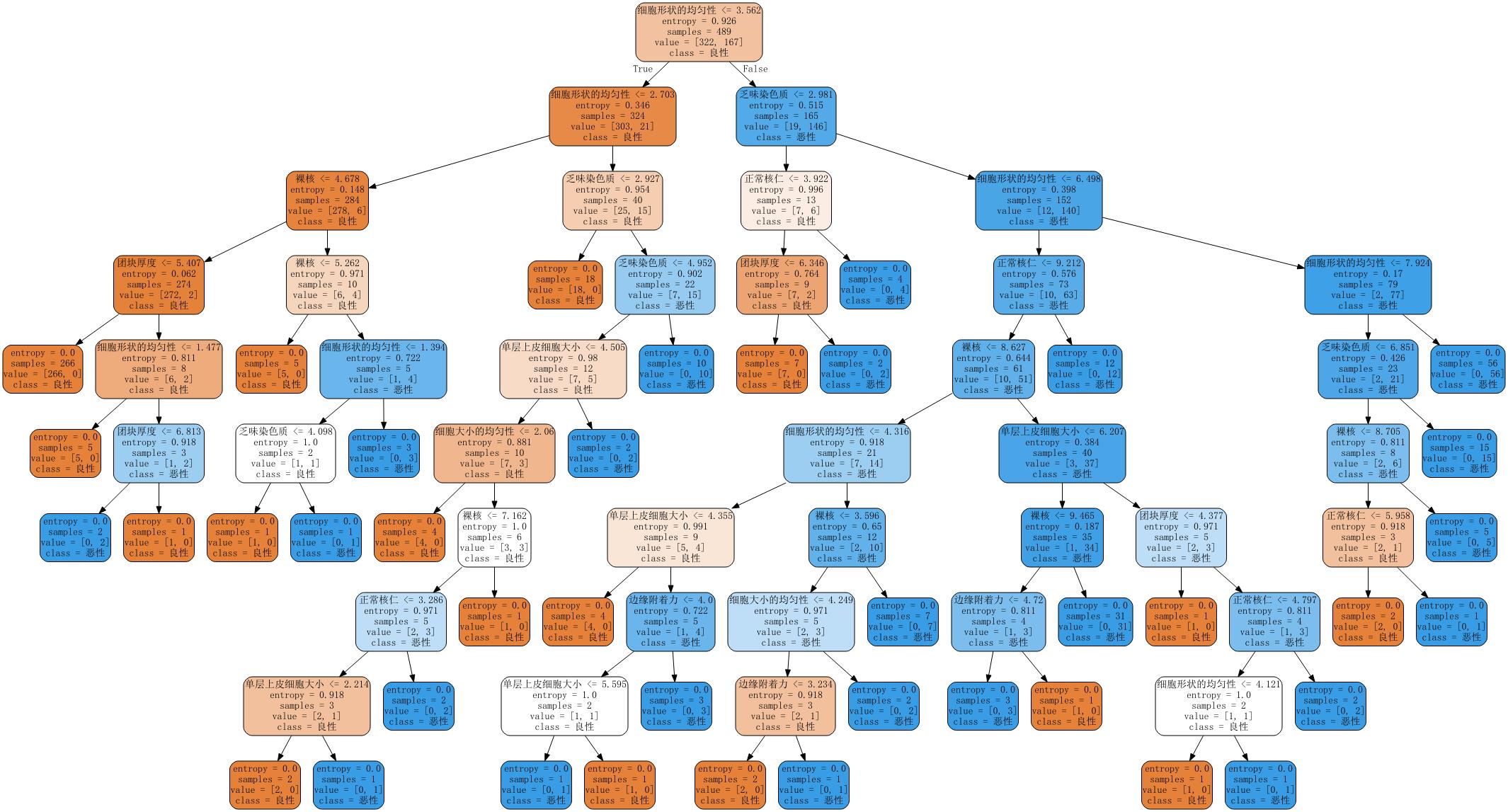
graphviz画图在这里有说:sklearn_决策树
特征重要性
[*zip(feature_name,clf.feature_importances_)]
[(‘团块厚度’, 0.07928484090157319),
(‘细胞大小的均匀性’, 0.006239695181160565),
(‘细胞形状的均匀性’, 0.6418075641411535),
(‘边缘附着力’, 0.025559378816469004),
(‘单层上皮细胞大小’, 0.012259788451959396),
(‘裸核’, 0.14909304212842098),
(‘乏味染色质’, 0.018736153376255508),
(‘正常核仁’, 0.06214238979422003),
(‘线粒体’, 0.004877147208787858)]
调参
默认参数
sklearn.tree.DecisionTreeClassifier(
criterion=’gini’,
splitter=’best’,
max_depth=None,
min_samples_split=2,
min_samples_leaf=1,
min_weight_fraction_leaf=0.0,
max_features=None,
random_state=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
class_weight=None,
presort=False)
可以进行网格搜索
# 用GridSearchCV寻找最优参数(字典)
# 导入库
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import GridSearchCV
param = 'criterion':['gini','entropy'],'max_depth':[ int(x) for x in list(np.linspace(1,10,10)) ],'min_samples_leaf':[2,3,4,5,10],'min_impurity_decrease':[0.1,0.2,0.5]
grid = GridSearchCV(DecisionTreeClassifier(),param_grid=param,cv=10)
grid.fit(Xtrain, Ytrain)
print('最优分类器:',grid.best_params_,'最优分数:', grid.best_score_) # 得到最优的参数和分值
最优分类器: ‘criterion’: ‘gini’, ‘max_depth’: 1, ‘min_impurity_decrease’: 0.1, ‘min_samples_leaf’: 2 最优分数: 0.9088860544217686
我做出来 效果不ok
- max_depth
#限制树的最大深度,超过设定深度的树枝全部剪掉
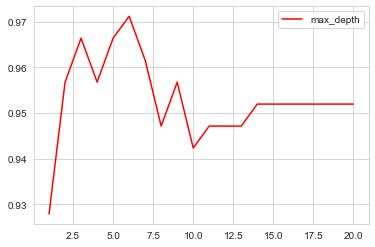
import matplotlib.pyplot as plt
test = []
for i in range(20):
clf = tree.DecisionTreeClassifier(max_depth=i+1
,criterion="entropy"
,random_state=30
,splitter="random")
clf = clf.fit(Xtrain, Ytrain)
score = clf.score(Xtest, Ytest)
test.append(score)
plt.plot(range(1,21),test,color="red",label="max_depth")
plt.legend()
plt.show()
print(max(test))
print(test.index(max(test))+1)
# 在 max_depth=6 时,得分最高
clf = tree.DecisionTreeClassifier(max_depth=6
,criterion="entropy"
,random_state=30
,splitter="random")
clf = clf.fit(Xtrain, Ytrain)
score = clf.score(Xtest, Ytest)
score
0.9711538461538461
tr = []
te = []
for i in range(10):
clf = DecisionTreeClassifier(random_state=25,max_depth=i+1,criterion="entropy")
clf = clf.fit(Xtrain, Ytrain)
score_tr = clf.score(Xtrain,Ytrain)
score_te = cross_val_score(clf,X,y,cv=10).mean()
tr.append(score_tr)
te.append(score_te)
print(max(te))
plt.plot(range(1,11),tr,color="red",label="train")
plt.plot(range(1,11),te,color="blue",label="test")
plt.xticks(range(1,11))
plt.legend()
plt.show()

明显的过拟合
- min_samples_leaf
限定一个节点在分枝后的每个子节点都必须包含至少min_samples_leaf个训练样本,否则分枝就不会发生
test = []
for i in range(10):
clf = tree.DecisionTreeClassifier(min_samples_leaf=i+1
,criterion="entropy"
,max_depth=6
,random_state=30
,splitter="random")
clf = clf.fit(Xtrain, Ytrain)
score = clf.score(Xtest, Ytest)
test.append(score)
plt.plot(range(1,11),test,color="blue",label="min_samples_leaf")
plt.legend()
plt.show()
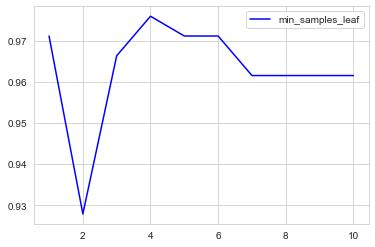
clf = tree.DecisionTreeClassifier(max_depth=6
,criterion="entropy"
,random_state=30
,splitter="random",min_samples_leaf=4)
clf = clf.fit(Xtrain, Ytrain)
score = clf.score(Xtest, Ytest)
score
0.9759615384615384
- min_samples_split
限定,一个节点必须要包含至少min_samples_split个训练样本,这个节点才允许被分枝,否则分枝就不会发生。
test = []
for i in range(9):
clf = tree.DecisionTreeClassifier(min_samples_split=i+2
,criterion="entropy"
,max_depth=6
,random_state=30
,splitter="random",min_samples_leaf=4)
clf = clf.fit(Xtrain, Ytrain)
score = clf.score(Xtest, Ytest)
test.append(score)
plt.plot(range(2,11),test,color="red",label="min_samples_split")
plt.legend()
plt.show()

clf = tree.DecisionTreeClassifier(max_depth=6
,criterion="entropy"
,random_state=30
,splitter="random",min_samples_leaf=4,min_samples_split=2)
clf = clf.fit(Xtrain, Ytrain)
score = clf.score(Xtest, Ytest)
score
0.9759615384615384
y_predict=clf.predict(Xtest)# 预测
y_predict

# 依然使用sklearn.metrics里面的classification_report模块对预测结果做更加详细的分析。
from sklearn.metrics import classification_report
print (classification_report(Ytest, y_predict))
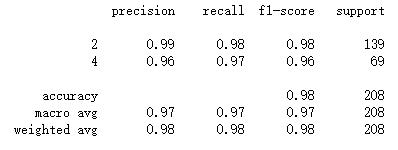
支持向量机
from sklearn.svm import SVC
clf = SVC(kernel = "rbf"
,gamma=以上是关于案例:乳腺癌威斯康星州(原始)数据集的主要内容,如果未能解决你的问题,请参考以下文章