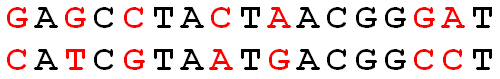06 Counting Point Mutations
Posted thinkanddo
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了06 Counting Point Mutations相关的知识,希望对你有一定的参考价值。
Problem
Given two strings ss and tt of equal length, the Hamming distance between ss and tt, denoted dH(s,t)dH(s,t), is the number of corresponding symbols that differ in ss and tt. See Figure 2.
Given: Two DNA strings ss and tt of equal length (not exceeding 1 kbp).
Return: The Hamming distance dH(s,t)dH(s,t).
Sample Dataset
GAGCCTACTAACGGGAT CATCGTAATGACGGCCT
Sample Output
7
方法一:
s = ‘GAGCCTACTAACGGGAT‘
t = ‘CATCGTAATGACGGCCT‘
count = 0
for i in xrange(len(s)):
if s[i] != t[i]:
count +=1
print count
方法二:
### Counting Point Mutations ###
fh = open(‘/Users/DONG/Downloads/rosalind_hamm.txt‘, ‘rt‘)
seq = fh.readlines()
a,b = seq[0].strip(),seq[1].strip()
hammingDistance = 0
for i in range(len(a)):
if a[i] != b[i]:
hammingDistance += 1
print (hammingDistance)
以上是关于06 Counting Point Mutations的主要内容,如果未能解决你的问题,请参考以下文章