pymol-note
Posted jszd
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了pymol-note相关的知识,希望对你有一定的参考价值。
一、Snapshot(用不同颜色显示蛋白不同二级结构,用不同颜色显示不同小分子,显示连键)
>load name.pdb,name
#>fetch object
#>disable object-name
#>enable object-name
#>delete selection-name
#>show representation #其中representation可以为:cartoon, ribbon, dots, spheres, surface和mesh
#>hide representation
#>as representation
如下:
>load last.pdb,2fkw
>hide all
>show cartoon,2fkw
>dss 2fkw
>color red,ss h
>color yellow,ss s
>color blue,ss “”
> set cartoon_transparency, 0.5
>select bcl,resn BCL
>show sticks,bcl
>color green,bcl
>select rg1,resn RG1
>show sticks,rg1
>color orange,rg1
>set stick_radius,0.15 #将小分子显示为球棍模型
>set sphere_scale, 0.2
>show spheres, bcl
>show spheres, rg1
>select mg,name MG
>color pink,mg
>alter mg,vdw=2
>rebuild
>wizard measurement #显示原子之间距离:或者wizard-measurement
>hide labels
二、加载namd动力学轨迹:
首先将dcd文件用cpptraj转化为mdcrd文件,然后再pymol中运行命令
加载轨迹:
>load prmtop,mmm,format=top
>load_traj 02_Heat.mdcrd,mmm,format=trj, start=1, stop=10001, interval=100
>hide
>show cartoon,mmm
>dss mmm #计算二级结构
> color red,ss h
> color blue,ss l+
> color yellow,ss s
>select THM,selection
>color yellow,THM
>show sticks,THM
>bg_color white
>ray 900,900
三、动画
# 定义动画
mset 1 x30
# mdo命令创建摇摆+/-180度的30帧动画
util.mrock start, finish, angle, phase, loop-flag
util.mrock 1,30,180,1,1
mplay
>bg_color white
>ray
>save test.png

**********
pymol>zoom all
>color red,all
> hide
> dss 1kim
> show cartoon,1kim
> color red,ss h
> color blue,ss l+
> color yellow,ss s
>select THM,selection
>color yellow,THM
>show sticks,THM
>bg_color white
>ray 900,900

Cited from: http://www.sohu.com/a/218617791_777125
Pymol> log_open script-file-name.pml #记录一个文本文档,该文件的后缀名应为.pml
Pymol> log_close # 终止记录
Pymol> @script-file-name # 调用该文档
外部GUI窗口里面的File - Save Session,创建一个会话文件(.pse),下次打开Pymol时直接回到当前所在的状态。
Pymol> png file-name # 图片被保存在PyMOL安装默认的文件夹中
pymol> fetch 1w2i # 载入一个pdb分子
Hide everything and then show protein cartoon
• PyMOL> hide everything, all
• PyMOL> show cartoon, all
Color the helix, sheet, and loop
• PyMOL> color purple, ss h
• PyMOL> color yellow, ss s
• PyMOL> color green, ss ""
Color chain A and B
• PyMOL> color red, chain A
• PyMOL> color blue, chain B
Color the active site residue
PyMOL> select active, (resi 14-20,38 and chain A)
PyMOL> color yellow, active
PyMOL> turn y, -60; turn x, -20
PyMOL> zoom active
Locate and display the bound formate ion in the active site.
PyMOL> select ligand, active around 3.5 and resn FMT
PyMOL> show sticks, ligand
PyMOL> show spheres, ligand
PyMOL> alter ligand, vdw=0.5
PyMOL> rebuild
PyMOL> set transparency=0.25
8Rendering and output
• PyMOL> bg_color white
• PyMOL> ray
• File -> Save Image
Display the side-chain of active site residues on top of the cartoon representation
• PyMOL> hide surface
• PyMOL> select sidechain, not (name c+n+o)
• PyMOL> show sticks, (active and sidechain)
• PyMOL> color blue, name n*
• PyMOL> color red, name o*
• PyMOL> color white, name c*
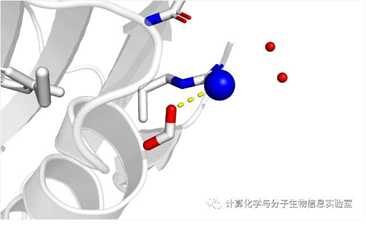
接下来进行距离的衡量,Wizard -> Measurement -> Distance,进行稍微的美化,把cartoon的透明度调整后,即可得到下图:
以上是关于pymol-note的主要内容,如果未能解决你的问题,请参考以下文章