Introduction to Writing Functions in R
Posted gaowenxingxing
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了Introduction to Writing Functions in R相关的知识,希望对你有一定的参考价值。
在R中编写函数
为啥要用函数
为了代码更加可读,减少重复代码==lazy
You can type less code, saving effort and making your analyses more readable.
You can reuse your code from project to project.
in R function are a type of variable ,so you assign functions using left arrow,just like anything else
args(函数名)
查询函数的参数
> args(rank)
function (x, na.last = TRUE, ties.method = c("average", "first",
"last", "random", "max", "min")) 创建一个函数的步骤
# Update the function so heads have probability p_head
toss_coin <- function(n_flips,p_head) {
coin_sides <- c("head", "tail")
# Define a vector of weights
weights <- c(p_head,1-p_head)
# Modify the sampling to be weighted
sample(coin_sides, n_flips, replace = TRUE,prob=weights)
}
# Generate 10 coin tosses
toss_coin(10,0.8)think of function as verbs
1.default args
在定义函数的时候就已经赋值了
cut_by_quantile <- function(x, n=5, na.rm, labels, interval_type) {
probs <- seq(0, 1, length.out = n + 1)
qtiles <- quantile(x, probs, na.rm = na.rm, names = FALSE)
right <- switch(interval_type, "(lo, hi]" = TRUE, "[lo, hi)" = FALSE)
cut(x, qtiles, labels = labels, right = right, include.lowest = TRUE)
}
# Remove the n argument from the call
cut_by_quantile(
n_visits,
na.rm = FALSE,
labels = c("very low", "low", "medium", "high", "very high"),
interval_type = "(lo, hi]"
)
n是默认参数若默认参数存在了,那么调用的时候只给形参传值好了
# Set the categories for interval_type to "(lo, hi]" and "[lo, hi)"
cut_by_quantile <- function(x, n = 5, na.rm = FALSE, labels = NULL,
interval_type = c("(lo, hi]", "[lo, hi)")) {
# Match the interval_type argument
interval_type <- match.arg(interval_type)
probs <- seq(0, 1, length.out = n + 1)
qtiles <- quantile(x, probs, na.rm = na.rm, names = FALSE)
right <- switch(interval_type, "(lo, hi]" = TRUE, "[lo, hi)" = FALSE)
cut(x, qtiles, labels = labels, right = right, include.lowest = TRUE)
}
# Remove the interval_type argument from the call
cut_by_quantile(n_visits)Passing arguments between functions
# From previous steps
get_reciprocal <- function(x) {
1 / x
}
calc_harmonic_mean <- function(x) {
x %>%
get_reciprocal() %>%
mean() %>%
get_reciprocal()
}
std_and_poor500 %>%
# Group by sector
group_by(sector) %>%
# Summarize, calculating harmonic mean of P/E ratio
summarize(hmean_pe_ratio = calc_harmonic_mean(pe_ratio))calc_harmonic_mean <- function(x, ...) {
x %>%
get_reciprocal() %>%
mean(...) %>%
get_reciprocal()
}
>
> std_and_poor500 %>%
# Group by sector
group_by(sector) %>%
# Summarize, calculating harmonic mean of P/E ratio
summarize(hmean_pe_ratio = calc_harmonic_mean(pe_ratio,na.rm=TRUE))
# A tibble: 11 x 2
sector hmean_pe_ratio
<chr> <dbl>
1 Communication Services 17.5
2 Consumer Discretionary 15.2
3 Consumer Staples 19.8
4 Energy 13.7
5 Financials 12.9
6 Health Care 26.6
7 Industrials 18.2
8 Information Technology 21.6
9 Materials 16.3
10 Real Estate 32.5
11 Utilities 23.9Checking arguments
You can use the assert_*() functions from assertive to check inputs and throw errors when they fail.
calc_harmonic_mean <- function(x, na.rm = FALSE) {
# Assert that x is numeric
assert_is_numeric(x)
x %>%
get_reciprocal() %>%
mean(na.rm = na.rm) %>%
get_reciprocal()
}弹出报错
calc_harmonic_mean <- function(x, na.rm = FALSE) {
assert_is_numeric(x)
# Check if any values of x are non-positive
if(any(is_non_positive(x), na.rm = TRUE)) {
# Throw an error
stop("x contains non-positive values, so the harmonic mean makes no sense.")
}
x %>%
get_reciprocal() %>%
mean(na.rm = na.rm) %>%
get_reciprocal()
}
# See what happens when you pass it negative numbers
calc_harmonic_mean(std_and_poor500$pe_ratio - 20)固定函数参数
一直存在的且默认
Returning values from functions
Sometimes, you don‘t need to run through the whole body of a function to get the answer. In that case you can return early from that function using return().
To check if x is divisible by n, you can use is_divisible_by(x, n) from assertive.
Alternatively, use the modulo operator, %%. x %% n gives the remainder when dividing x by n, so x %% n == 0 determines whether x is divisible by n. Try 1:10 %% 3 == 0 in the console.
To solve this exercise, you need to know that a leap year is every 400th year (like the year 2000) or every 4th year that isn‘t a century (like 1904 but not 1900 or 1905).
is_leap_year <- function(year) {
# If year is div. by 400 return TRUE
if(is_divisible_by(year, 400)) {
return(TRUE)
}
# If year is div. by 100 return FALSE
if(is_divisible_by(year, 100)) {
return(FALSE)
}
# If year is div. by 4 return TRUE
if(is_divisible_by(year, 4)) {
return(TRUE)
}
# Otherwise return FALSE
FALSE
}# Define a pipeable plot fn with data and formula args
pipeable_plot <- function(data, formula) {
# Call plot() with the formula interface
plot(formula, data)
# Invisibly return the input dataset
invisible(data)
}
# Draw the scatter plot of dist vs. speed again
plt_dist_vs_speed <- cars %>%
pipeable_plot(dist ~ speed)
# Now the plot object has a value
plt_dist_vs_speedaugments
Given an R statistical model or other non-tidy object, add columns to the original dataset such as predictions, residuals and cluster assignments.
> # Use broom tools to get a list of 3 data frames
> list(
# Get model-level values
model = glance(model),
# Get coefficient-level values
coefficients = tidy(model),
# Get observation-level values
observations = augment(model)
)
$model
# A tibble: 1 x 7
null.deviance df.null logLik AIC BIC deviance df.residual
<dbl> <int> <dbl> <dbl> <dbl> <dbl> <int>
1 18850. 345 -6425. 12864. 12891. 11529. 339
$coefficients
# A tibble: 7 x 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 4.09 0.0279 146. 0.
2 genderfemale 0.374 0.0212 17.6 2.18e- 69
3 income($25k,$55k] -0.0199 0.0267 -0.746 4.56e- 1
4 income($55k,$95k] -0.581 0.0343 -16.9 3.28e- 64
5 income($95k,$Inf) -0.578 0.0310 -18.7 6.88e- 78
6 travel(0.25h,4h] -0.627 0.0217 -28.8 5.40e-183
7 travel(4h,Infh) -2.42 0.0492 -49.3 0.
$observations
# A tibble: 346 x 12
.rownames n_visits gender income travel .fitted .se.fit .resid .hat
<chr> <dbl> <fct> <fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 25 2 female ($95k~ (4h,I~ 1.46 0.0502 -1.24 0.0109
2 26 1 female ($95k~ (4h,I~ 1.46 0.0502 -1.92 0.0109
3 27 1 male ($95k~ (0.25~ 2.88 0.0269 -5.28 0.0129
4 29 1 male ($95k~ (4h,I~ 1.09 0.0490 -1.32 0.00711
5 30 1 female ($55k~ (4h,I~ 1.46 0.0531 -1.92 0.0121
6 31 1 male [$0,$~ [0h,0~ 4.09 0.0279 -10.4 0.0465
7 33 80 female [$0,$~ [0h,0~ 4.46 0.0235 -0.710 0.0479
8 34 104 female ($95k~ [0h,0~ 3.88 0.0261 6.90 0.0332
9 35 55 male ($25k~ (0.25~ 3.44 0.0222 3.85 0.0153
10 36 350 female ($25k~ [0h,0~ 4.44 0.0206 21.5 0.0360
# ... with 336 more rows, and 3 more variables: .sigma <dbl>, .cooksd <dbl>,
# .std.resid <dbl>Magnificent multi-assignment! Returning many values is as easy as collecting them into a list. The groomed model has data frames that are easy to program against.
# From previous step
groom_model <- function(model) {
list(
model = glance(model),
coefficients = tidy(model),
observations = augment(model)
)
}
# Call groom_model on model, assigning to 3 variables
c(mdl, cff, obs) %<-% groom_model(model)
# See these individual variables
mdl; cff; obsEnvironments
> # From previous step
> rsa_lst <- list(
capitals = capitals,
national_parks = national_parks,
population = population
)
>
> # Convert the list to an environment
> rsa_env <- list2env(rsa_lst )
>
> # List the structure of each variable
> ls.str(rsa_env)
capitals : Classes 'tbl_df', 'tbl' and 'data.frame': 3 obs. of 2 variables:
$ city : chr "Cape Town" "Bloemfontein" "Pretoria"
$ type_of_capital: chr "Legislative" "Judicial" "Administrative"
national_parks : chr [1:22] "Addo Elephant National Park" "Agulhas National Park" ...
population : Time-Series [1:5] from 1996 to 2016: 40583573 44819778 47390900 51770560 55908900查看父类的环境变量
> # From previous steps
> rsa_lst <- list(
capitals = capitals,
national_parks = national_parks,
population = population
)
> rsa_env <- list2env(rsa_lst)
>
> # Find the parent environment of rsa_env
> parent <- parent.env(rsa_env)
>
> # Print its name
> environmentName(parent )
[1] "R_GlobalEnv"exists
查看是否定义了一个变量exists packages
> # Does population exist in rsa_env?
> exists("population", envir = rsa_env)
[1] TRUE
>
> # Does population exist in rsa_env, ignoring inheritance?
> exists("population", envir = rsa_env,inherits=FALSE)
[1] FALSE局部变量和全局变量 这个解释可以参考之前的python学习笔记python 函数学习笔记
这里个人觉得编程语言是在某种程度上面是相通的,还是认真的学习好一门吧。不然最后都会放弃了,加油
单位换算
# Write a function to convert sq. meters to hectares
sq_meters_to_hectares <- function(sq_meters) {
sq_meters / 10000
}
# Write a function to convert acres to sq. yards
acres_to_sq_yards <- function(acres) {
4840 * acres
}
# Write a function to convert acres to sq. yards
acres_to_sq_yards <- function(acres) {
4840 * acres
}常见农业单位换算
There are 4840 square yards in an acre.
There are 36 inches in a yard and one inch is 0.0254 meters.
There are 10000 square meters in a hectare.
# Write a function to convert sq. yards to sq. meters
sq_yards_to_sq_meters <- function(sq_yards) {
sq_yards %>%
# Take the square root
sqrt() %>%
# Convert yards to meters
yards_to_meters() %>%
# Square it
raise_to_power(2)
}单位换算有可以直接用的函数
# Load fns defined in previous steps
load_step3()
# Write a function to convert bushels to kg
bushels_to_kgs <- function(bushels, crop) {
bushels %>%
# Convert bushels to lbs for this crop
bushels_to_lbs(crop) %>%
# Convert lbs to kgs
lbs_to_kgs()
}# Wrap this code into a function
fortify_with_metric_units <- function(data, crop) {
data %>%
mutate(
farmed_area_ha = acres_to_hectares(farmed_area_acres),
yield_kg_per_ha = bushels_per_acre_to_kgs_per_hectare(
yield_bushels_per_acre,
crop = crop
)
)
}
# Try it on the wheat dataset
fortify_with_metric_units(wheat, crop = "wheat")绘图
# Using corn, plot yield (kg/ha) vs. year
> ggplot(corn, aes(year, yield_kg_per_ha)) +
# Add a line layer, grouped by state
geom_line(aes(group = state)) +
# Add a smooth trend layer
geom_smooth()inner_join()
# Wrap this code into a function
fortify_with_census_region<-function(data){
data %>%
inner_join(usa_census_regions, by = "state")
}
# Try it on the wheat dataset
fortify_with_census_region(wheat)# Wrap this code into a function
plot_yield_vs_year_by_region<-function(data){
plot_yield_vs_year(data) +
facet_wrap(vars(census_region))
}
# Try it on the wheat dataset
plot_yield_vs_year_by_region(wheat)Modeling grain yields
# Wrap the model code into a function
run_gam_yield_vs_year_by_region<-function(data){
gam(yield_kg_per_ha ~ s(year) + census_region, data = corn)
}
# Try it on the wheat dataset
run_gam_yield_vs_year_by_region(wheat)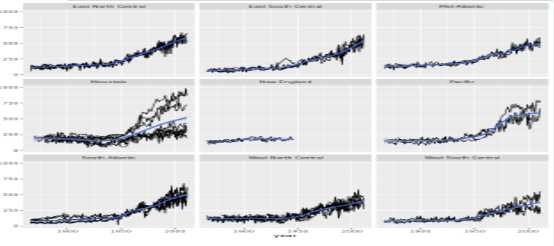
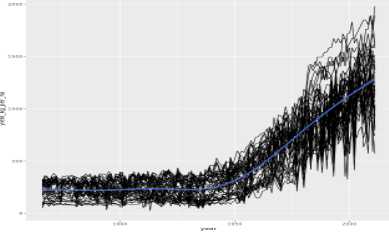
# Make predictions in 2050
predict_this <- data.frame(
year = 2050,
census_region = census_regions
)
# Predict the yield
pred_yield_kg_per_ha <- predict(corn_model, predict_this, type = "response")
predict_this %>%
# Add the prediction as a column of predict_this
mutate(pred_yield_kg_per_ha = pred_yield_kg_per_ha)reference
以上是关于Introduction to Writing Functions in R的主要内容,如果未能解决你的问题,请参考以下文章
redis -Writing to master: Unknown error异常处理
redis -Writing to master: Unknown error异常处理
Lecture3 Structure of Academic Writing
FlinkFlink 报错 Writing records to streamload failed