文献导读 | Single-Cell Sequencing of iPSC-Dopamine Neurons Reconstructs Disease Progression and Identifi
Posted leezx
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了文献导读 | Single-Cell Sequencing of iPSC-Dopamine Neurons Reconstructs Disease Progression and Identifi相关的知识,希望对你有一定的参考价值。
目前唯一一篇把单细胞测序应用到疾病发育上的高分paper,发表在了CSC上。
参考阅读:
首先明确一下概念:
帕金森疾病有一定的遗传因素,本文就是研究可遗传的这部分,GBA基因里的N370S突变会导致帕金森。The majority of PD cases are idiopathic, with only about 10% attributed to inherited PD cases
早期发现:increased ER stress, autophagic and lysosomal perturbations, and elevated a-synuclein release;内质网应激增加,自噬和溶酶体微扰增加,a-synuclein释放增加
基本的思路:
找出了一个核心的DEG:HDAC4
HDAC4靶向药物可以纠正帕金森症状
把核心DEG按照作用时间进行了分类。
根据临床信息把不同的疾病样本分层了stratify
HDAC4错误定位了 mislocalized
问题:
1. 如何找出HDAC4的?
这并不是一个转录因子
Pseudotime analysis of genes differentially expressed (DE) along this axis identified the transcriptional repressor histone deacetylase 4 (HDAC4) as an upstream regulator of disease progression.
HDAC4 was mislocalized to the nucleus in PD iPSC-derived dopamine neurons and repressed genes early in the disease axis, leading to late deficits in protein homeostasis.
HDAC4错误地进入了细胞核,抑制了那些早起基因,导致了蛋白质稳态缺陷。
2. 怎么找到靶向药物的?
3. 如何构建pseudotime的?
we identified a progressive axis of gene expression variation leading to endoplasmic reticulum stress.
Combining bulk and single-cell expression profiles, we identified a robust set of 60 genes whose expression captured an axis of variation between cells from controls and the remaining two PD GBA-N370S patients.
4. core gene set是怎么找出来的?
We next sought to identify a core set of genes consistently perturbed across both bulk RNA-seq and the single-cell transcriptomic signature. This core set was identified as the intersection of two gene sets from the analysis: (1) those DE in bulk RNAseq using DESeq2 after the removal of GBA3 (at 5% FDR) and (2) those DE across PC2 using switchde (at 5% FDR; Campbell and Yau, 2017; Figure S4B). We further refined this set to include only those additionally identified as discriminating marker genes after clustering the single-cell RNA-seq data using SC3 (Kiselev et al., 2017). By combining genes found through both bulk and single-cell DE as well as single-cell clustering (STAR Methods), we identified a core set of 60 genes, 52 of which were consistently downregulated and 8 of which were consistently upregulated in PD GBA-N370S iPSC-derived dopamine neurons (Figure S4B).
去文中一一找答案。。。
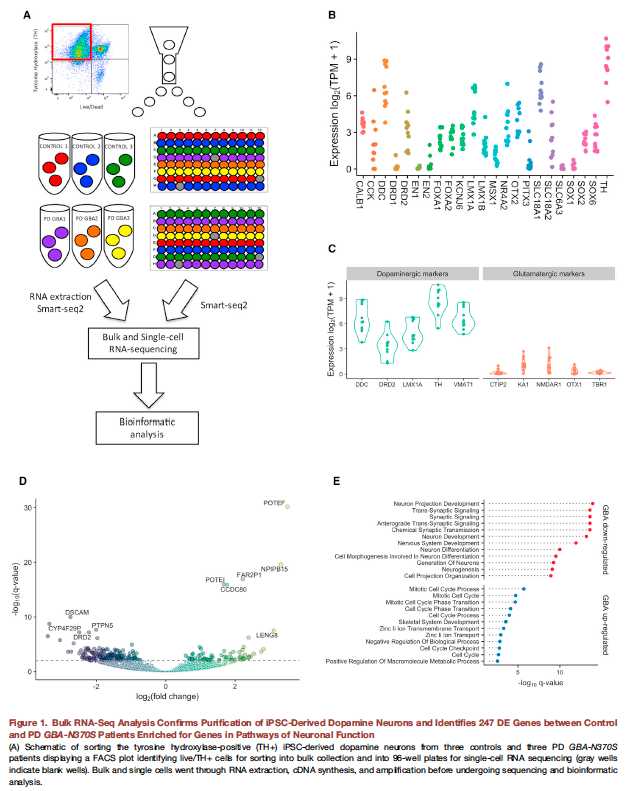
图1:
A:提纯purify iPSC-derived dopamine neurons,然后做bulk RNA-seq 和单细胞
B:increased expression of dopamine neuron marker genes多巴胺神经元的marker表达提升。感觉图表很业余,看得费力,大致就是想展现一些marker的表达情况,证明细胞是对的;
C:常规DEG的火山图,对比的bulk RNA-seq,我们不能用,因为样本不够纯;
D:常规的GO 注释图,down and up分类
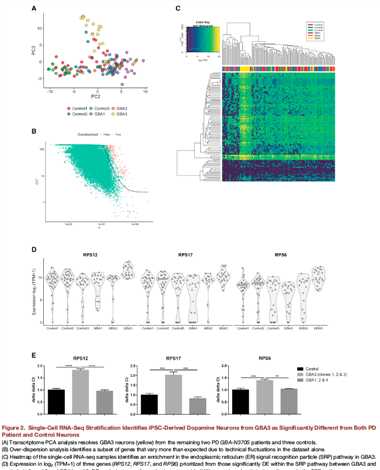
图二:
A:单细胞常规的聚类分析,只不过用得是PC2和PC3,应该用了scater包,发现了分层现象,GBA3的表达变异被PC3捕获了;
B:常规的Over dispersion分析,鉴定出具有生物学差异的基因;才143个gene,有没有搞错;
C:an enrichment in the endoplasmic reticulum (ER) signal recognition particle (SRP) pathway in GBA3;ER通路在GBA3中高度富集;
D:挑了几个基因出来证明;
E:qRTPCR验证,靶向膜途径的SRP依赖性共翻译蛋白的升高对GBA3具有特异性,提出了本研究中使用的患者的分子分层,分子分层的依据;
The stratification of PSP from PD among these GBAN370S carriers by single-cell profiling of their iPSC-derived dopamine neurons is therefore consistent with the clinical stratification and reveals a potential disease-relevant pathway for PSP.
我们的这个项目适合分层吗?肯定可以
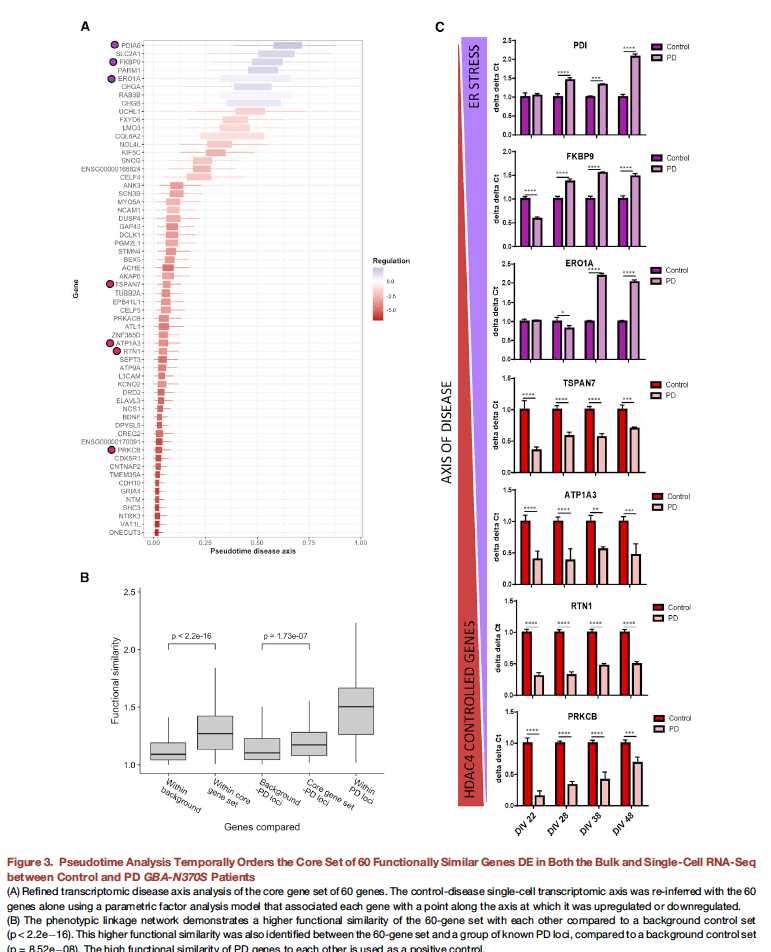
图三:
A:首先想办法构建了一个pseudo diease axis,然后判断core gene set在哪个位置发挥作用
B:统计学验证了core gene set的合理性;
C:挑了几个基因做了验证,DIV 22就是分化的天数,可以直观的看出基因到底是在哪一个区段开始发挥作用
以上是关于文献导读 | Single-Cell Sequencing of iPSC-Dopamine Neurons Reconstructs Disease Progression and Identifi的主要内容,如果未能解决你的问题,请参考以下文章