基于Sklearn的乳腺癌检测问题
Posted irivingloveluna
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了基于Sklearn的乳腺癌检测问题相关的知识,希望对你有一定的参考价值。
乳腺癌检测问题
1.使用数据:sklearn自带乳腺癌数据集
- data shape:(569, 30)
- no.postive:357个阳性
- no.negative:212个阴性
- features name:[‘mean radius‘ ‘mean texture‘ ‘mean perimeter‘ ‘mean area‘
‘mean smoothness‘ ‘mean compactness‘ ‘mean concavity‘
‘mean concave points‘ ‘mean symmetry‘ ‘mean fractal dimension‘
‘radius error‘ ‘texture error‘ ‘perimeter error‘ ‘area error‘
‘smoothness error‘ ‘compactness error‘ ‘concavity error‘
‘concave points error‘ ‘symmetry error‘ ‘fractal dimension error‘
‘worst radius‘ ‘worst texture‘ ‘worst perimeter‘ ‘worst area‘
‘worst smoothness‘ ‘worst compactness‘ ‘worst concavity‘
‘worst concave points‘ ‘worst symmetry‘ ‘worst fractal dimension‘]
1 #载入数据 2 from sklearn.datasets import load_breast_cancer 3 from sklearn.linear_model import LogisticRegression 4 from sklearn.model_selection import train_test_split 5 from sklearn.model_selection import cross_val_predict 6 from sklearn.model_selection import ShuffleSplit 7 from sklearn.preprocessing import PolynomialFeatures 8 from sklearn.model_selection import learning_curve 9 from sklearn.model_selection import ShuffleSplit 10 from sklearn.pipeline import Pipeline 11 import numpy as np 12 import matplotlib.pyplot as plt 13 import time 14 from common.utils import plot_learning_curve 15 16 %pylab 17 # np.random.seed(6) 18 19 #增加多项式做预处理 20 def polynomial_model(degree=1,**kwarg): 21 polynomial_features = PolynomialFeatures(degree=degree, 22 include_bias=False) 23 logistic_regression = LogisticRegression(**kwarg) 24 pipeline = Pipeline([("polynomial_features",polynomial_features), 25 ("logistic_regression",logistic_regression)]) 26 return pipeline 27 28 # 学习曲线的绘制 29 def plot_learning_curve(plt,estimator,title,X,y,ylim=None,cv=None, 30 n_jobs=1,train_sizes=np.linspace(.1,1.0,5)): 31 """ 32 estimator:实现‘fit‘和‘predict‘方法的对象类型 33 title:字符串:图标抬头 34 X,y:图标x,y 35 ylim:元祖,(ymin,ymax),可选择定义图像的最小和最大y值 36 cv:int,交叉验证生成器,可选择和决定交叉验证的生成策略,cv的可能输入有: 37 - None,使用默认的3-fold交叉验证 38 - Integer,指定折叠的数量 39 --如实上述两种情况,如果‘y‘是个二元的或者多元分类,使用StratifiedKFold 40 --如果‘y‘不是分类问题,使用KFold 41 - 一个对象,可被用来作为cv生成器 42 - 一个迭代器 43 n_jobs:整数,同时运行的工作数量 44 train_sizes:表示把训练样本数量从0.1~1分成5等分,生成[0.1,0.325,0.55,0.775,1]的 45 序列,从序列中取出训练样本数量百分比,逐个计算在当前训练样本数量情况下训练出来的 46 模型准确性。 47 """ 48 plt.title(title) 49 if ylim is not None: 50 plt.ylim(*ylim) 51 plt.xlabel("Training examples") 52 plt.ylabel("Score") 53 train_sizes,train_scores,test_scores=learning_curve(estimator, 54 X,y,cv=cv,n_jobs=n_jobs,train_sizes=train_sizes) 55 train_scores_mean = np.mean(train_scores,axis = 1) 56 train_scores_std = np.std(train_scores,axis = 1) 57 test_scores_mean = np.mean(test_scores,axis = 1) 58 test_scores_std = np.std(test_scores,axis = 1) 59 plt.grid(True) 60 plt.fill_between(train_sizes, train_scores_mean - train_scores_std, 61 train_scores_mean + train_scores_std, alpha=0.1, 62 color="r") 63 plt.fill_between(train_sizes,test_scores_mean-test_scores_std, 64 test_scores_mean+test_scores_std,alpha=0.1,color=‘g‘) 65 plt.plot(train_sizes,train_scores_mean,‘o--‘,color="r", 66 label = "Training score") 67 plt.plot(train_sizes,test_scores_mean,‘o-‘,color="g", 68 label = "Cross_Validation score") 69 plt.legend(loc="best") 70 return plt 71 72 cancer = load_breast_cancer() 73 X = cancer.data 74 y = cancer.target 75 print(‘data shape:{0}‘.format(X.shape)) 76 print(‘no.features:{0}‘.format(X.shape[1])) 77 print(‘features name:{0}‘.format(cancer.feature_names)) 78 print(‘no.postive:{0}‘.format(y[y==1].shape[0])) 79 print(‘no.negative:{0}‘.format(y[y==0].shape[0])) 80 # print(cancer.data[0]) 81 82 #模型训练 训练集和测试集 83 X_train,X_test,y_train,y_test = train_test_split(X,y,test_size = 0.2) 84 85 #模型训练 86 model = LogisticRegression() 87 model.fit(X_train,y_train) 88 train_score = model.score(X_train,y_train) 89 test_score = model.score(X_test,y_test) 90 print(‘ train score:{}‘.format(train_score)) 91 print(‘test score:{}‘.format(test_score)) 92 93 #样本预测 94 y_pred = model.predict(X_test) 95 print(‘ matchs:{0}/{1}‘.format(np.equal(y_pred,y_test).shape[0],y_test.shape[0])) 96 97 #预测概率 找出预测“自信度”低于90%的样本 阳性,阴性均大于0.1 98 y_pred_proba = model.predict_proba(X_test) 99 #找出第一列预测为阴性的概率大于0.1的样本,保存在result中 100 y_pred_proba_0 = y_pred_proba[:,0]>0.1 101 result = y_pred_proba[y_pred_proba_0] 102 #找出第二列预测为阳性的概率大于0.1的样本 103 y_pred_prob_1 = result[:,1]>0.1 104 print(result[y_pred_prob_1]) 105 106 # np.random.seed(3) 107 #模型优化 108 X_train1,X_test1,y_train1,y_test1 = train_test_split(X,y,test_size = 0.2) 109 model_adjust = polynomial_model(degree=2,penalty = ‘l1‘) 110 start = time.clock() 111 model_adjust.fit(X_train1,y_train1) 112 train_score1 = model.score(X_train1,y_train1) 113 cv_score = model.score(X_test1,y_test1) 114 print(‘ train score:{0:.6f}‘.format(train_score1)) 115 print(‘test score:{0:.6f}‘.format(cv_score)) 116 logistic_regression = model_adjust.named_steps[‘logistic_regression‘] 117 #coef_里面保存的是模型参数,增加二阶多项式后,特征由30-->495个,其实有用的只有94个 118 print(‘ model parameters shape:{0}‘.format(logistic_regression.coef_.shape)) 119 print(‘count of non-zero element:{0}‘.format(np.count_nonzero(logistic_regression.coef_))) 120 121 #首先画出L1作为正则项的一二三阶多项式学习曲线 122 cv = ShuffleSplit(n_splits=10,test_size=0.2,random_state=0) 123 title = ‘Learning_Curves(degree={0},penalty={1})‘ 124 degrees = [1,2,3] 125 penalty = [‘l1‘,‘l2‘] 126 index_penalty = 0 127 index_degrees = 0 128 count = 0 129 plt.figure(figsize=(15,4),dpi=100) 130 for i in range(6): 131 plt.subplot(2,len(degrees),i+1) 132 plot_learning_curve(plt,polynomial_model(degree=degrees[index_degrees],133 penalty=penalty[index_penalty]),134 title.format(degrees[index_degrees],penalty[index_penalty]),135 X,y,ylim=(0.8,1.01),cv=cv) 136 if i==len(degrees)-1: 137 index_penalty+=1 138 index_degrees=-1 139 index_degrees+=1 140 141 142 143 144
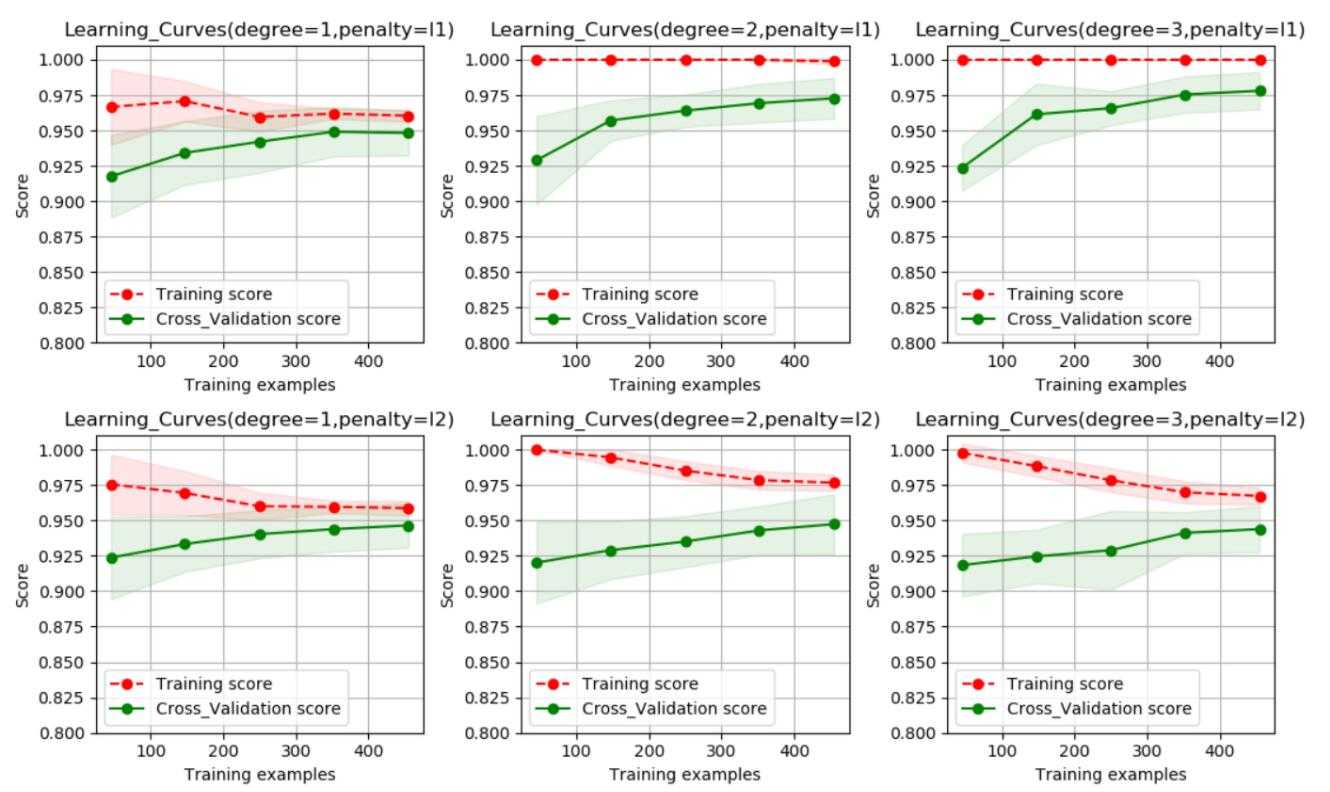
1.可以看出使用二阶段像是并使用L1范数作为正则项的模型为最优模型;
2.可以看出,训练样本评分和交叉验证样本评分之间的间隙还比较大,即方差比较大,可以采集更多的数据,以便于对模型进行优化。
以上是关于基于Sklearn的乳腺癌检测问题的主要内容,如果未能解决你的问题,请参考以下文章