手写体数字图像聚类实验代码怎么写
Posted
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了手写体数字图像聚类实验代码怎么写相关的知识,希望对你有一定的参考价值。
参考技术A 本文所有实现代码均来自《Python机器学习及实战》#-*- coding:utf-8 -*-
#分别导入numpy、matplotlib、pandas,用于数学运算、作图以及数据分析
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
#第一步:使用pandas读取训练数据和测试数据
digits_train = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/optdigits/optdigits.tra',header=None)
digits_test = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/optdigits/optdigits.tes',header=None)
#第二步:已知原始数据有65个特征值,前64个是像素特征,最后一个是每个图像样本的数字类别
#从训练集和测试集上都分离出64维度的像素特征和1维度的数字目标
X_train = digits_train[np.arange(64)]
y_train = digits_train[64]
X_test = digits_test[np.arange(64)]
y_test = digits_test[64]
#第三步:使用KMeans模型进行训练并预测
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=10)
kmeans.fit(X_train)
kmeans_y_predict = kmeans.predict(X_test)
#第四步:评估KMeans模型的性能
#如何评估聚类算法的性能?
#1.Adjusted Rand Index(ARI) 适用于被用来评估的数据本身带有正确类别的信息,ARI指标和计算Accuracy的方法类似
#2.Silhouette Coefficient(轮廓系数) 适用于被用来评估的数据没有所属类别 同时兼顾了凝聚度和分散度,取值范围[-1,1],值越大,聚类效果越好
from sklearn.metrics import adjusted_rand_score
print 'The ARI value of KMeans is',adjusted_rand_score(y_test,kmeans_y_predict)
#到此为止,手写体数字图像聚类--kmeans学习结束,下面单独讨论轮廓系数评价kmeans的性能
#****************************************************************************************************
#拓展学习:利用轮廓系数评价不同累簇数量(k值)的K-means聚类实例
from sklearn.metrics import silhouette_score
#分割出3*2=6个子图,并且在1号子图作图 subplot(nrows, ncols, plot_number)
plt.subplot(3,2,1)
#初始化原始数据点
x1 = np.array([1,2,3,1,5,6,5,5,6,7,8,9,7,9])
x2 = np.array([1,3,2,2,8,6,7,6,7,1,2,1,1,3])
# a = [1,2,3] b = [4,5,6] zipped = zip(a,b) 输出为元组的列表[(1, 4), (2, 5), (3, 6)]
X = np.array(zip(x1,x2)).reshape(len(x1),2)
#X输出为:array([[1, 1],[2, 3],[3, 2],[1, 2],...,[9, 3]])
#在1号子图作出原始数据点阵的分布
plt.xlim([0,10])
plt.ylim([0,10])
plt.title('Instances')
plt.scatter(x1,x2)
colors = ['b','g','r','c','m','y','k','b']
markers = ['o','s','D','v','^','p','*','+']
clusters = [2,3,4,5,8]
subplot_counter = 1
sc_scores = []
for t in clusters:
subplot_counter += 1
plt.subplot(3,2,subplot_counter)
kmeans_model = KMeans(n_clusters=t).fit(X)
for i,l in enumerate(kmeans_model.labels_):
plt.plot(x1[i],x2[i],color=colors[l],marker=markers[l],ls='None')
plt.xlim([0,10])
plt.ylim([0,10])
sc_score = silhouette_score(X,kmeans_model.labels_,metric='euclidean')
sc_scores.append(sc_score)
#绘制轮廓系数与不同类簇数量的直观显示图
plt.title('K=%s,silhouette coefficient = %0.03f'%(t,sc_score))
#绘制轮廓系数与不同类簇数量的关系曲线
plt.figure() #此处必须空一行,表示在for循环结束之后执行!!!
plt.plot(clusters,sc_scores,'*-') #绘制折线图时的样子
plt.xlabel('Number of Clusters')
plt.ylabel('Silhouette Coefficient Score')
plt.show()
#****************************************************************************************************
#总结:
#k-means聚类模型所采用的迭代式算法,直观易懂并且非常实用,但是有两大缺陷
#1.容易收敛到局部最优解,受随机初始聚类中心影响,可多执行几次k-means来挑选性能最佳的结果
#2.需要预先设定簇的数量,
深度学习实验:Softmax实现手写数字识别
文章相关知识点:AI遮天传 DL-回归与分类_老师我作业忘带了的博客-CSDN博客
MNIST数据集
MNIST手写数字数据集是机器学习领域中广泛使用的图像分类数据集。它包含60,000个训练样本和10,000个测试样本。这些数字已进行尺寸规格化,并在固定尺寸的图像中居中。每个样本都是一个784×1的矩阵,是从原始的28×28灰度图像转换而来的。MNIST中的数字范围是0到9。下面显示了一些示例。 注意:在训练期间,切勿以任何形式使用有关测试样本的信息。

代码清单
- data/ 文件夹:存放MNIST数据集。下载数据,解压后存放于该文件夹下。下载链接:MNIST handwritten digit database, Yann LeCun, Corinna Cortes and Chris Burges
- solver.py 这个文件中实现了训练和测试的流程。;
- dataloader.py 实现了数据加载器,可用于准备数据以进行训练和测试;
- visualize.py 实现了plot_loss_and_acc函数,该函数可用于绘制损失和准确率曲线;
- optimizer.py 实现带momentum的SGD优化器,可用于执行参数更新;
- loss.py 实现softmax_cross_entropy_loss,包含loss的计算和梯度计算;
- runner.ipynb 完成所有代码后的执行文件,执行训练和测试过程。
要求
- 记录训练和测试的准确率。画出训练损失和准确率曲线;
- 比较使用和不使用momentum结果的不同,可以从训练时间,收敛性和准确率等方面讨论差异;
- 调整其他超参数,如学习率,Batchsize等,观察这些超参数如何影响分类性能。写下观察结果并将这些新结果记录在报告中。
运行结果如下:


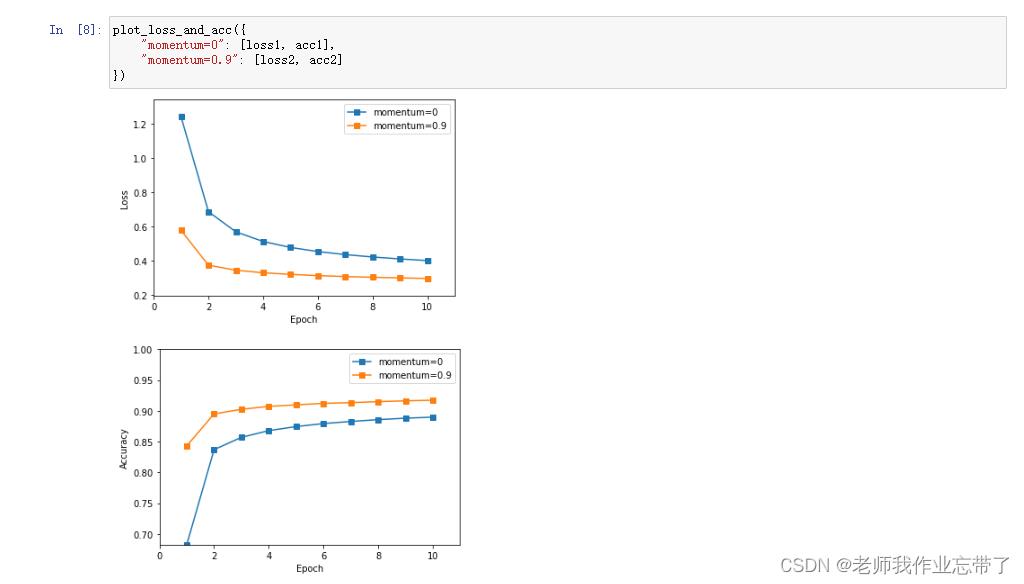
代码如下:
solver.py
import numpy as np
from layers import FCLayer
from dataloader import build_dataloader
from network import Network
from optimizer import SGD
from loss import SoftmaxCrossEntropyLoss
from visualize import plot_loss_and_acc
class Solver(object):
def __init__(self, cfg):
self.cfg = cfg
# build dataloader
train_loader, val_loader, test_loader = self.build_loader(cfg)
self.train_loader = train_loader
self.val_loader = val_loader
self.test_loader = test_loader
# build model
self.model = self.build_model(cfg)
# build optimizer
self.optimizer = self.build_optimizer(self.model, cfg)
# build evaluation criterion
self.criterion = SoftmaxCrossEntropyLoss()
@staticmethod
def build_loader(cfg):
train_loader = build_dataloader(
cfg['data_root'], cfg['max_epoch'], cfg['batch_size'], shuffle=True, mode='train')
val_loader = build_dataloader(
cfg['data_root'], 1, cfg['batch_size'], shuffle=False, mode='val')
test_loader = build_dataloader(
cfg['data_root'], 1, cfg['batch_size'], shuffle=False, mode='test')
return train_loader, val_loader, test_loader
@staticmethod
def build_model(cfg):
model = Network()
model.add(FCLayer(784, 10))
return model
@staticmethod
def build_optimizer(model, cfg):
return SGD(model, cfg['learning_rate'], cfg['momentum'])
def train(self):
max_epoch = self.cfg['max_epoch']
epoch_train_loss, epoch_train_acc = [], []
for epoch in range(max_epoch):
iteration_train_loss, iteration_train_acc = [], []
for iteration, (images, labels) in enumerate(self.train_loader):
# forward pass
logits = self.model.forward(images)
loss, acc = self.criterion.forward(logits, labels)
# backward_pass
delta = self.criterion.backward()
self.model.backward(delta)
# updata the model weights
self.optimizer.step()
# restore loss and accuracy
iteration_train_loss.append(loss)
iteration_train_acc.append(acc)
# display iteration training info
if iteration % self.cfg['display_freq'] == 0:
print("Epoch [][]\\t Batch [][]\\t Training Loss :.4f\\t Accuracy :.4f".format(
epoch, max_epoch, iteration, len(self.train_loader), loss, acc))
avg_train_loss, avg_train_acc = np.mean(iteration_train_loss), np.mean(iteration_train_acc)
epoch_train_loss.append(avg_train_loss)
epoch_train_acc.append(avg_train_acc)
# validate
avg_val_loss, avg_val_acc = self.validate()
# display epoch training info
print('\\nEpoch []\\t Average training loss :.4f\\t Average training accuracy :.4f'.format(
epoch, avg_train_loss, avg_train_acc))
# display epoch valiation info
print('Epoch []\\t Average validation loss :.4f\\t Average validation accuracy :.4f\\n'.format(
epoch, avg_val_loss, avg_val_acc))
return epoch_train_loss, epoch_train_acc
def validate(self):
logits_set, labels_set = [], []
for images, labels in self.val_loader:
logits = self.model.forward(images)
logits_set.append(logits)
labels_set.append(labels)
logits = np.concatenate(logits_set)
labels = np.concatenate(labels_set)
loss, acc = self.criterion.forward(logits, labels)
return loss, acc
def test(self):
logits_set, labels_set = [], []
for images, labels in self.test_loader:
logits = self.model.forward(images)
logits_set.append(logits)
labels_set.append(labels)
logits = np.concatenate(logits_set)
labels = np.concatenate(labels_set)
loss, acc = self.criterion.forward(logits, labels)
return loss, acc
if __name__ == '__main__':
# You can modify the hyerparameters by yourself.
relu_cfg =
'data_root': 'data',
'max_epoch': 10,
'batch_size': 100,
'learning_rate': 0.1,
'momentum': 0.9,
'display_freq': 50,
'activation_function': 'relu',
runner = Solver(relu_cfg)
relu_loss, relu_acc = runner.train()
test_loss, test_acc = runner.test()
print('Final test accuracy :.4f\\n'.format(test_acc))
# You can modify the hyerparameters by yourself.
sigmoid_cfg =
'data_root': 'data',
'max_epoch': 10,
'batch_size': 100,
'learning_rate': 0.1,
'momentum': 0.9,
'display_freq': 50,
'activation_function': 'sigmoid',
runner = Solver(sigmoid_cfg)
sigmoid_loss, sigmoid_acc = runner.train()
test_loss, test_acc = runner.test()
print('Final test accuracy :.4f\\n'.format(test_acc))
plot_loss_and_acc(
"relu": [relu_loss, relu_acc],
"sigmoid": [sigmoid_loss, sigmoid_acc],
)
dataloader.py
import os
import struct
import numpy as np
class Dataset(object):
def __init__(self, data_root, mode='train', num_classes=10):
assert mode in ['train', 'val', 'test']
# load images and labels
kind = 'train': 'train', 'val': 'train', 'test': 't10k'[mode]
labels_path = os.path.join(data_root, '-labels-idx1-ubyte'.format(kind))
images_path = os.path.join(data_root, '-images-idx3-ubyte'.format(kind))
with open(labels_path, 'rb') as lbpath:
magic, n = struct.unpack('>II', lbpath.read(8))
labels = np.fromfile(lbpath, dtype=np.uint8)
with open(images_path, 'rb') as imgpath:
magic, num, rows, cols = struct.unpack('>IIII', imgpath.read(16))
images = np.fromfile(imgpath, dtype=np.uint8).reshape(len(labels), 784)
if mode == 'train':
# training images and labels
self.images = images[:55000] # shape: (55000, 784)
self.labels = labels[:55000] # shape: (55000,)
elif mode == 'val':
# validation images and labels
self.images = images[55000:] # shape: (5000, 784)
self.labels = labels[55000:] # shape: (5000, )
else:
# test data
self.images = images # shape: (10000, 784)
self.labels = labels # shape: (10000, )
self.num_classes = 10
def __len__(self):
return len(self.images)
def __getitem__(self, idx):
image = self.images[idx]
label = self.labels[idx]
# Normalize from [0, 255.] to [0., 1.0], and then subtract by the mean value
image = image / 255.0
image = image - np.mean(image)
return image, label
class IterationBatchSampler(object):
def __init__(self, dataset, max_epoch, batch_size=2, shuffle=True):
self.dataset = dataset
self.batch_size = batch_size
self.shuffle = shuffle
def prepare_epoch_indices(self):
indices = np.arange(len(self.dataset))
if self.shuffle:
np.random.shuffle(indices)
num_iteration = len(indices) // self.batch_size + int(len(indices) % self.batch_size)
self.batch_indices = np.split(indices, num_iteration)
def __iter__(self):
return iter(self.batch_indices)
def __len__(self):
return len(self.batch_indices)
class Dataloader(object):
def __init__(self, dataset, sampler):
self.dataset = dataset
self.sampler = sampler
def __iter__(self):
self.sampler.prepare_epoch_indices()
for batch_indices in self.sampler:
batch_images = []
batch_labels = []
for idx in batch_indices:
img, label = self.dataset[idx]
batch_images.append(img)
batch_labels.append(label)
batch_images = np.stack(batch_images)
batch_labels = np.stack(batch_labels)
yield batch_images, batch_labels
def __len__(self):
return len(self.sampler)
def build_dataloader(data_root, max_epoch, batch_size, shuffle=False, mode='train'):
dataset = Dataset(data_root, mode)
sampler = IterationBatchSampler(dataset, max_epoch, batch_size, shuffle)
data_lodaer = Dataloader(dataset, sampler)
return data_lodaer
loss.py
import numpy as np
# a small number to prevent dividing by zero, maybe useful for you
EPS = 1e-11
class SoftmaxCrossEntropyLoss(object):
def forward(self, logits, labels):
"""
Inputs: (minibatch)
- logits: forward results from the last FCLayer, shape (batch_size, 10)
- labels: the ground truth label, shape (batch_size, )
"""
############################################################################
# TODO: Put your code here
# Calculate the average accuracy and loss over the minibatch
# Return the loss and acc, which will be used in solver.py
# Hint: Maybe you need to save some arrays for backward
self.one_hot_labels = np.zeros_like(logits)
self.one_hot_labels[np.arange(len(logits)), labels] = 1
self.prob = np.exp(logits) / (EPS + np.exp(logits).sum(axis=1, keepdims=True))
# calculate the accuracy
preds = np.argmax(self.prob, axis=1) # self.prob, not logits.
acc = np.mean(preds == labels)
# calculate the loss
loss = np.sum(-self.one_hot_labels * np.log(self.prob + EPS), axis=1)
loss = np.mean(loss)
############################################################################
return loss, acc
def backward(self):
############################################################################
# TODO: Put your code here
# Calculate and return the gradient (have the same shape as logits)
return self.prob - self.one_hot_labels
############################################################################
network.py
class Network(object):
def __init__(self):
self.layerList = []
self.numLayer = 0
def add(self, layer):
self.numLayer += 1
self.layerList.append(layer)
def forward(self, x):
# forward layer by layer
for i in range(self.numLayer):
x = self.layerList[i].forward(x)
return x
def backward(self, delta):
# backward layer by layer
for i in reversed(range(self.numLayer)): # reversed
delta = self.layerList[i].backward(delta)
optimizer.py
import numpy as np
class SGD(object):
def __init__(self, model, learning_rate, momentum=0.0):
self.model = model
self.learning_rate = learning_rate
self.momentum = momentum
def step(self):
"""One backpropagation step, update weights layer by layer"""
layers = self.model.layerList
for layer in layers:
if layer.trainable:
############################################################################
# TODO: Put your code here
# Calculate diff_W and diff_b using layer.grad_W and layer.grad_b.
# You need to add momentum to this.
# Weight update with momentum
if not hasattr(layer, 'diff_W'):
layer.diff_W = 0.0
layer.diff_W = layer.grad_W + self.momentum * layer.diff_W
layer.diff_b = layer.grad_b
layer.W += -self.learning_rate * layer.diff_W
layer.b += -self.learning_rate * layer.diff_b
# # Weight update without momentum
# layer.W += -self.learning_rate * layer.grad_W
# layer.b += -self.learning_rate * layer.grad_b
############################################################################
visualize.py
import matplotlib.pyplot as plt
import numpy as np
def plot_loss_and_acc(loss_and_acc_dict):
# visualize loss curve
plt.figure()
min_loss, max_loss = 100.0, 0.0
for key, (loss_list, acc_list) in loss_and_acc_dict.items():
min_loss = min(loss_list) if min(loss_list) < min_loss else min_loss
max_loss = max(loss_list) if max(loss_list) > max_loss else max_loss
num_epoch = len(loss_list)
plt.plot(range(1, 1 + num_epoch), loss_list, '-s', label=key)
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend()
plt.xticks(range(0, num_epoch + 1, 2))
plt.axis([0, num_epoch + 1, min_loss - 0.1, max_loss + 0.1])
plt.show()
# visualize acc curve
plt.figure()
min_acc, max_acc = 1.0, 0.0
for key, (loss_list, acc_list) in loss_and_acc_dict.items():
min_acc = min(acc_list) if min(acc_list) < min_acc else min_acc
max_acc = max(acc_list) if max(acc_list) > max_acc else max_acc
num_epoch = len(acc_list)
plt.plot(range(1, 1 + num_epoch), acc_list, '-s', label=key)
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.legend()
plt.xticks(range(0, num_epoch + 1, 2))
plt.axis([0, num_epoch + 1, min_acc, 1.0])
plt.show()
以上是关于手写体数字图像聚类实验代码怎么写的主要内容,如果未能解决你的问题,请参考以下文章