R语言颜色-图例legend及位置调整
Posted 生信小猪
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了R语言颜色-图例legend及位置调整相关的知识,希望对你有一定的参考价值。
R中的颜色设置
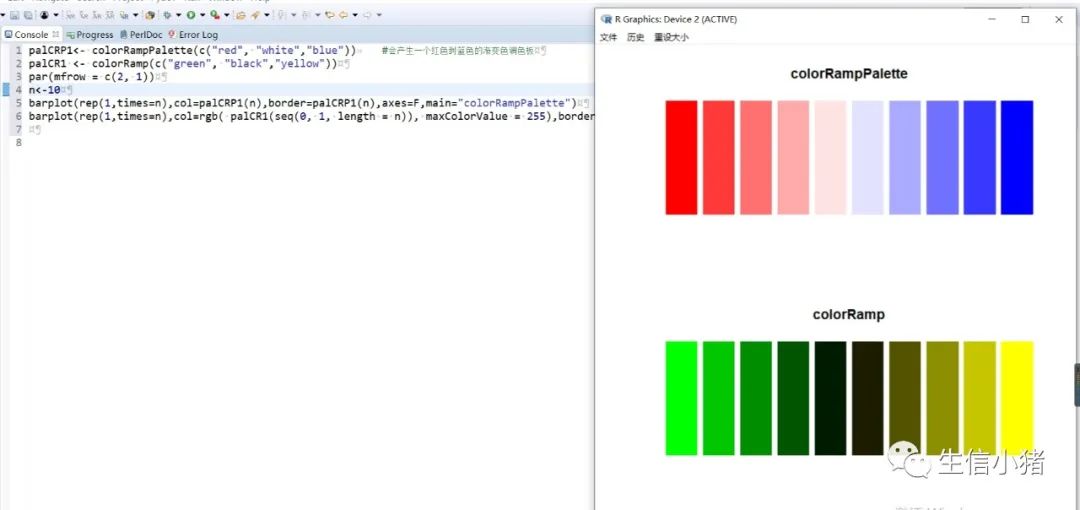
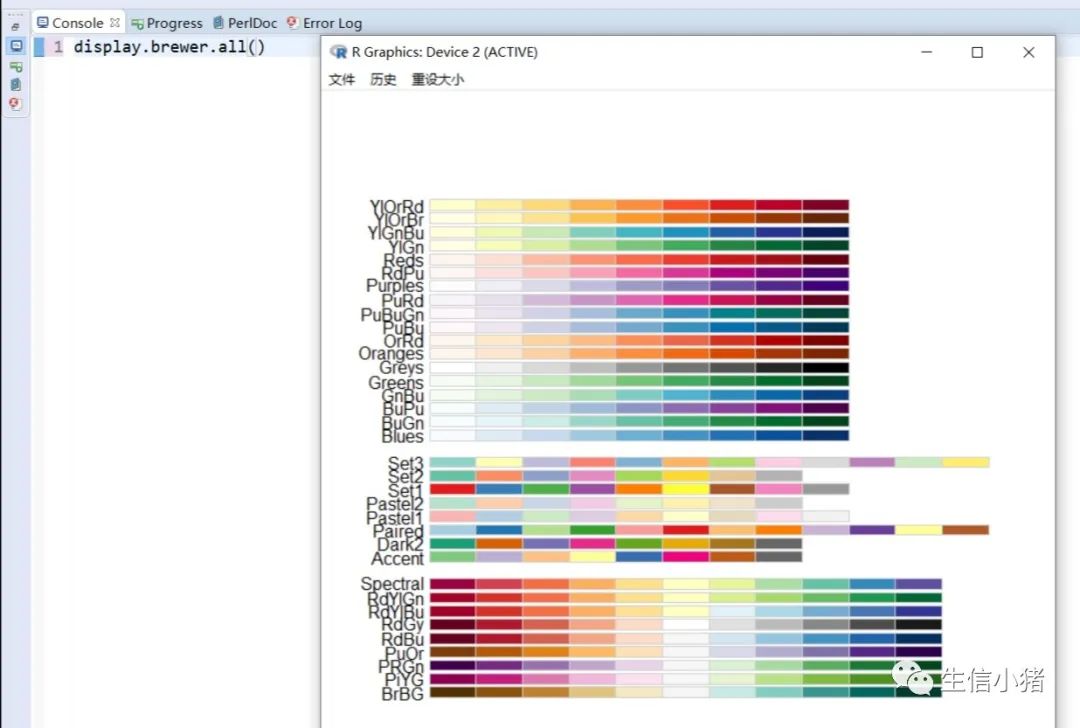
##################################R中颜色的表示palette() #查看色板,有8种颜色set.seed(323) #该命令的作用是设定生成随机数的种子,种子是为了让结果具有重复性。如果不设定种子,生成的随机数无法重现。plot(rnorm(4), col = c(1, "blue", rgb(0, 1, 0.1), "#FF0000"),pch=15,cex=2)#修改调色板palette(c("#377eb8","#4daf4a","#984ea3","#ff7f00","#ffff33","#a65628","#f781bf","#999999"))plot(rnorm(4), col = c(1, 2, 3, 4),pch=15,cex=2)#恢复调色板默认值palette("default")#透明颜色表示方法#rgb(1,0,0,0.5) "#FF000080" #在后面多加一个0.5的参数,是半透明;如果是16进制,加上80,就是8乘以16,就是128,相对于225也是一半,相当于半透明plot(rnorm(100),col=c(rgb(255,22,0,maxColorValue = 255),"#0000FF80"),pch=15,cex=2)#颜色名称对应颜色值col2rgb("blue")#################################R内置渐变色 调色板# rainbow()# heat.colors()# terrain.colors()# topo.colors()# cm.colors()#颜色展示:par(mfrow=c(5,1),mar=c(0.1,0.1,2,0.1),xaxs="i", yaxs="i") #xaxs="i"表示设置x轴的范围,默认值取“r”,表示坐标轴比给定作图范围(xlim给出的范围)稍微大一点儿,取”i”时表示坐标轴范围与给定作图范围完全相同。n<-10barplot(rep(1,times=n),col=rainbow(n),border=rainbow(n),axes=F,main="Rainbow Color") #barplot(rep(1,times=n),col=heat.colors(n),border=heat.colors(n),axes=F,main="Heat.Colors") #axes=F表示是否绘制坐标轴,默认是显示的barplot(rep(1,times=n),col=terrain.colors(n),border=terrain.colors(n),axes=F,main="Terrain.Colors")barplot(rep(1,times=n),col=topo.colors(n),border=topo.colors(n),axes=F,main="Topo.Colors")barplot(rep(1,times=n),col=cm.colors(n),border=cm.colors(n),axes=F,main="Cm.Colors")#用法,输入数字,返回颜色值rainbow(10)#################################自定义渐变色调色板:#colorRampPalette() 给出指定数量的颜色(将调色板等分)#colorRamp() 给一个0-1的数返回对应的颜色palCRP1<- colorRampPalette(c("red", "white","blue")) #会产生一个红色到蓝色的渐变色调色板palCR1 <- colorRamp(c("green", "black","yellow"))par(mfrow = c(2, 1))n<-1000barplot(rep(1,times=n),col=palCRP1(n),border=palCRP1(n),axes=F,main="colorRampPalette")barplot(rep(1,times=n),col=rgb( palCR1(seq(0, 1, length = n)), maxColorValue = 255),border=rgb( palCR1(seq(0, 1, length = n)), maxColorValue = 255),axes=F,main="colorRamp")#颜色搭配包:RColorBrewer,调色板#install.packages("RColorBrewer")require("RColorBrewer") #载入需要的程辑包:RColorBrewerdisplay.brewer.all() #显示所有颜色,这里有三种颜色方案brewer.pal(3, "Set1") #获取颜色:第一个参数表示调取颜色的数量,第二个参数表示调取颜色条为Set1的颜色方案
R语言中颜色表示方法
颜色数值表示法
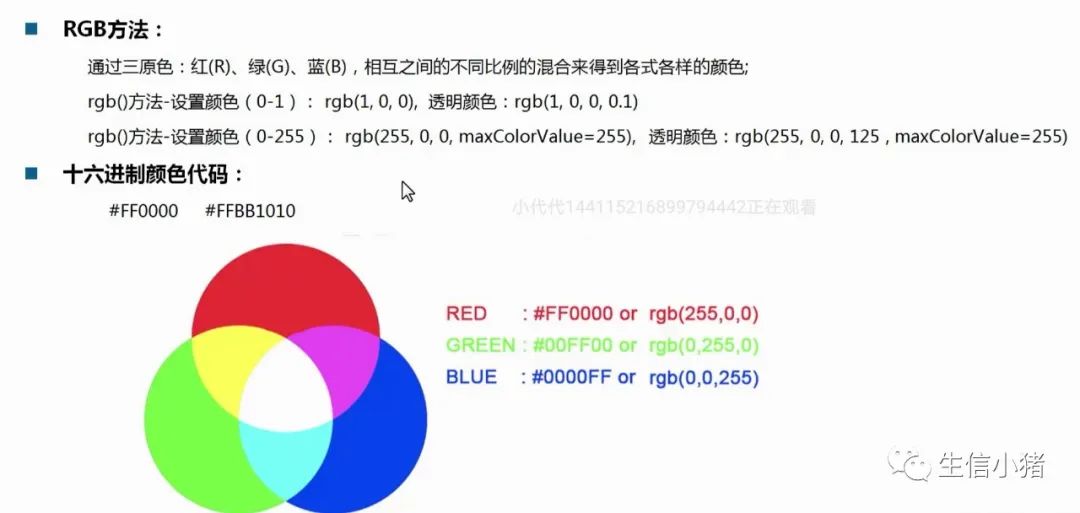
#FF00006位字母分别表示:FF表示红色,00表示绿色,00表示蓝色

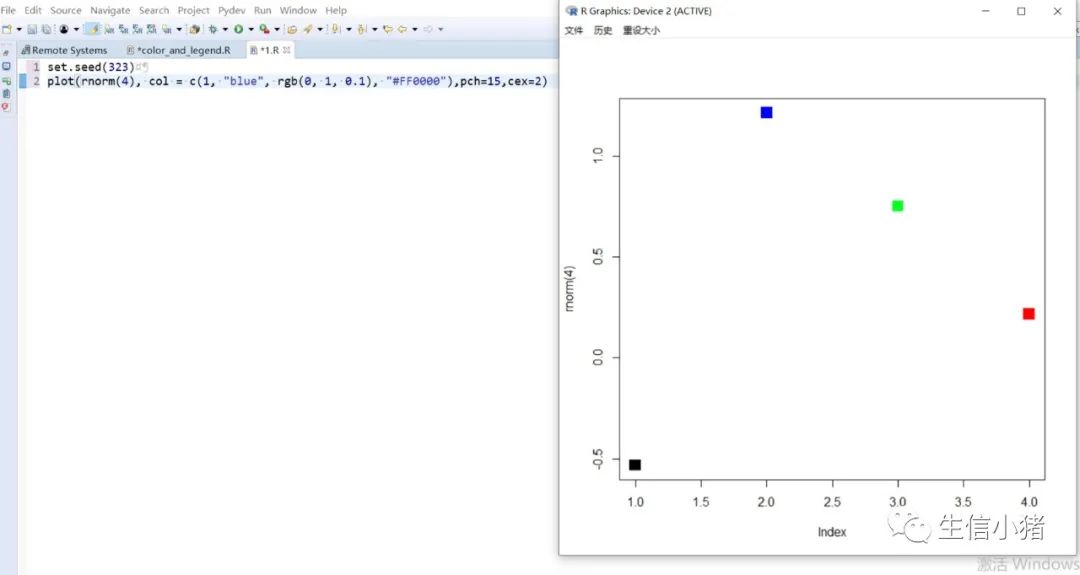
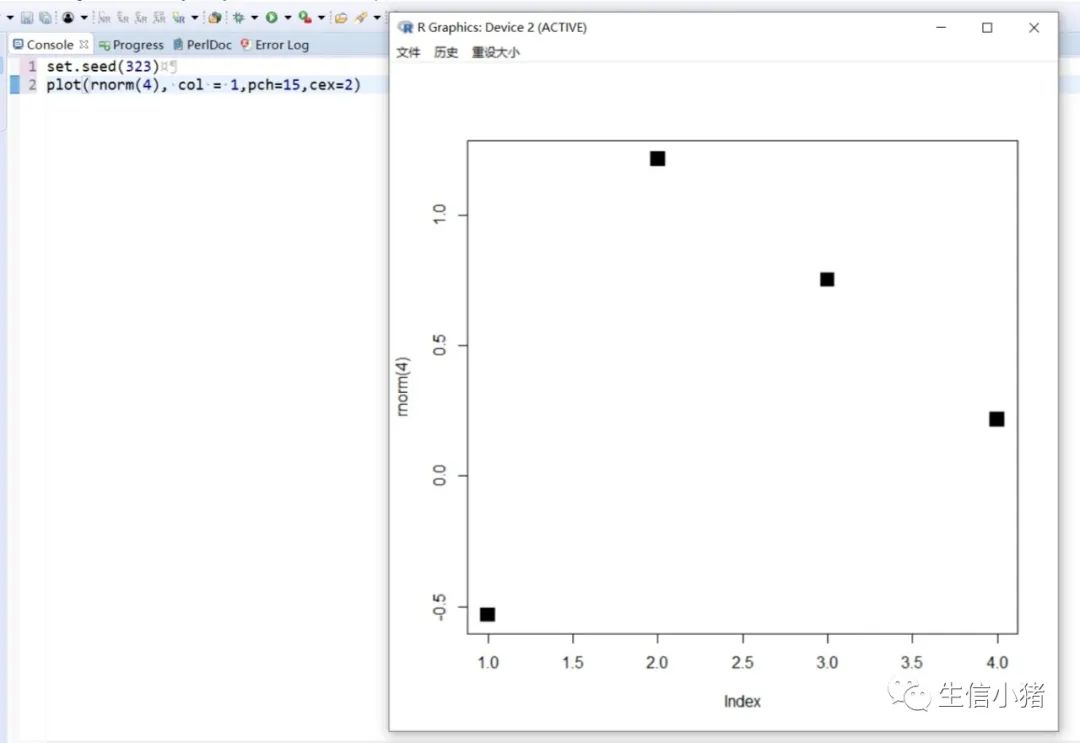
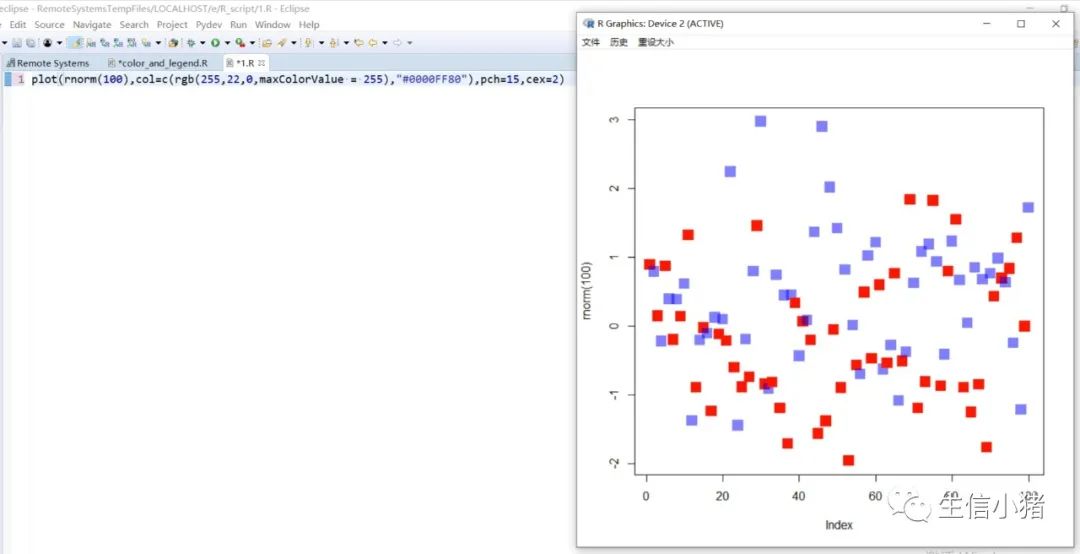

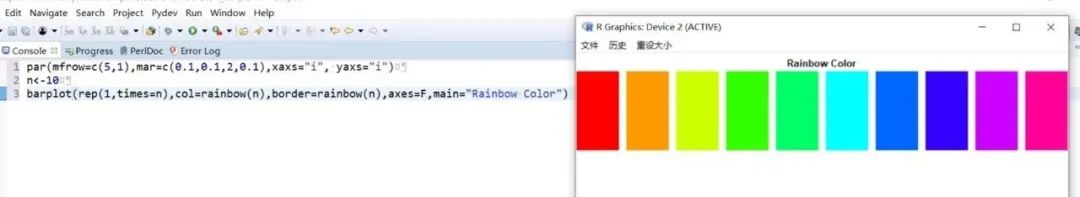
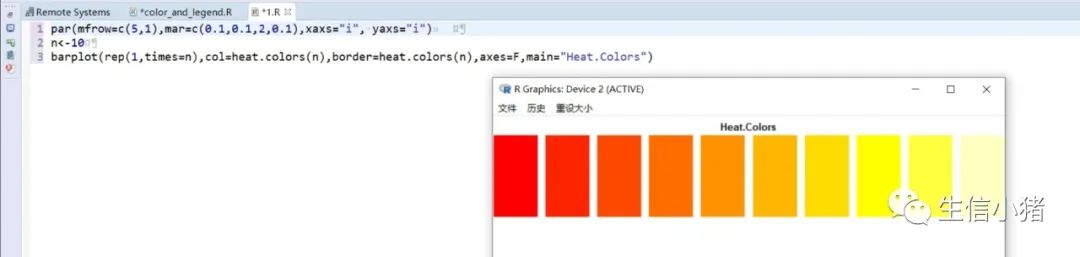
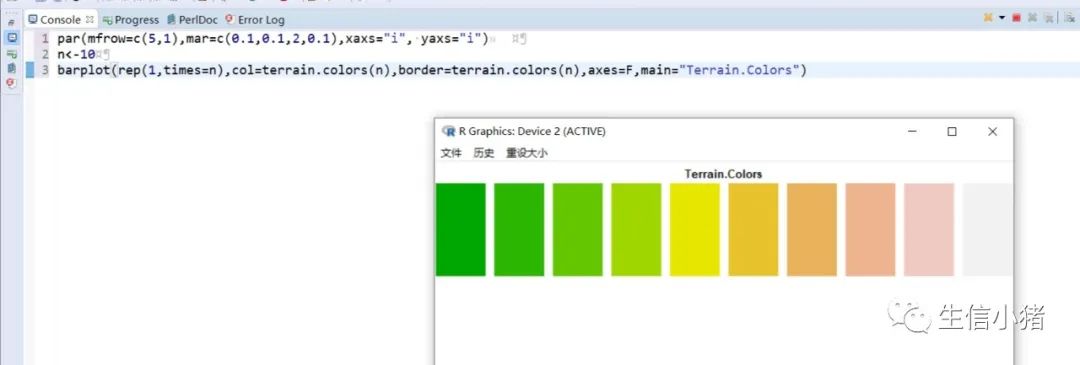
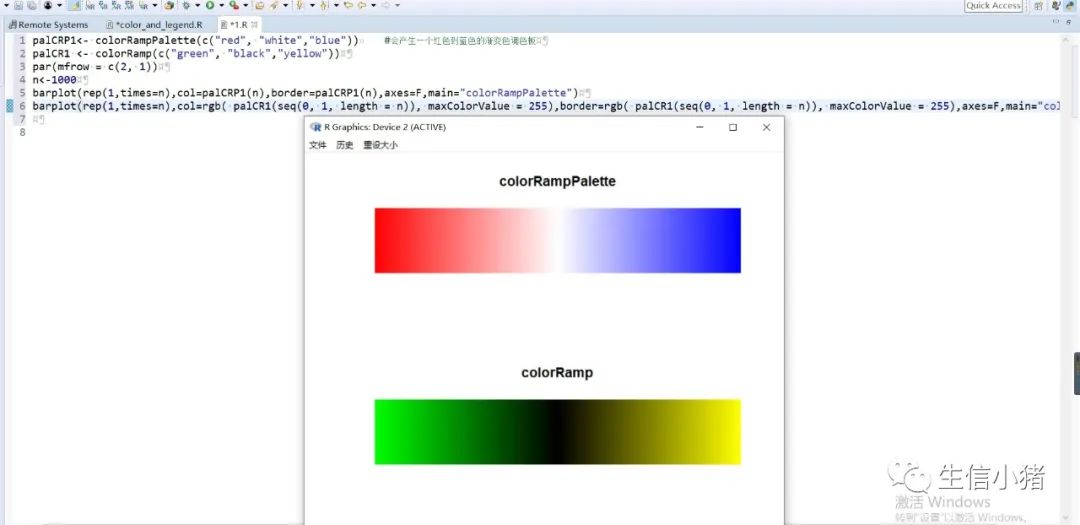
R中的图例设置
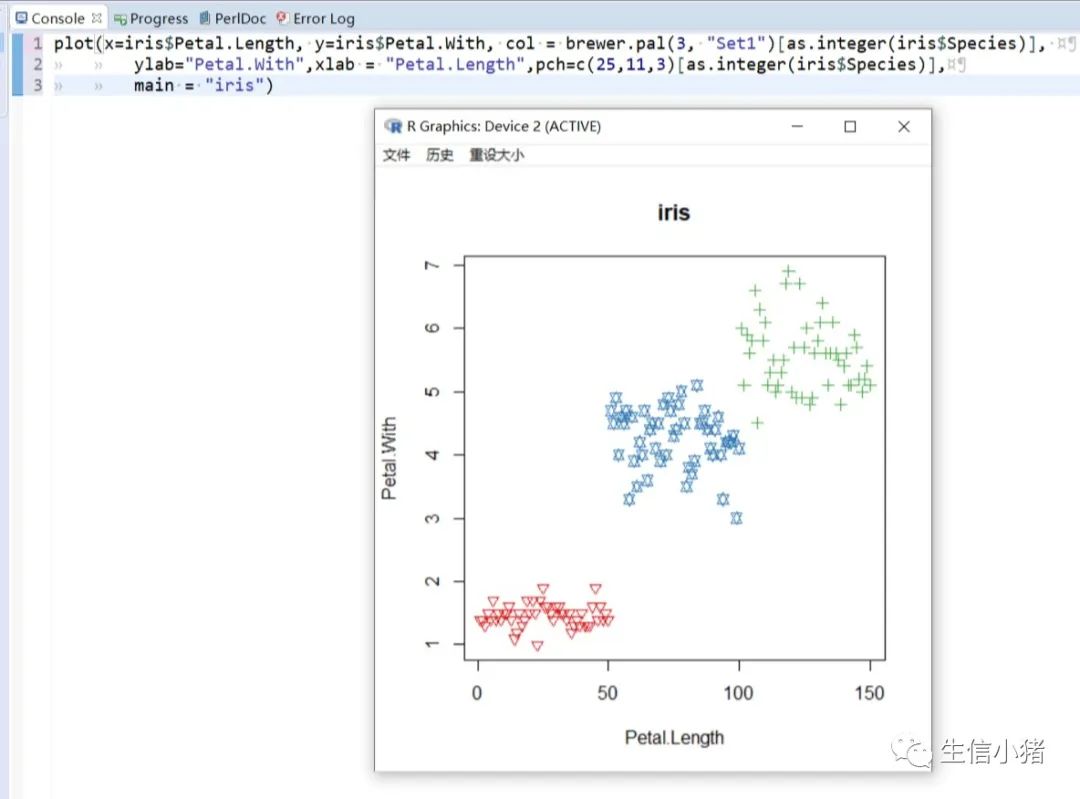
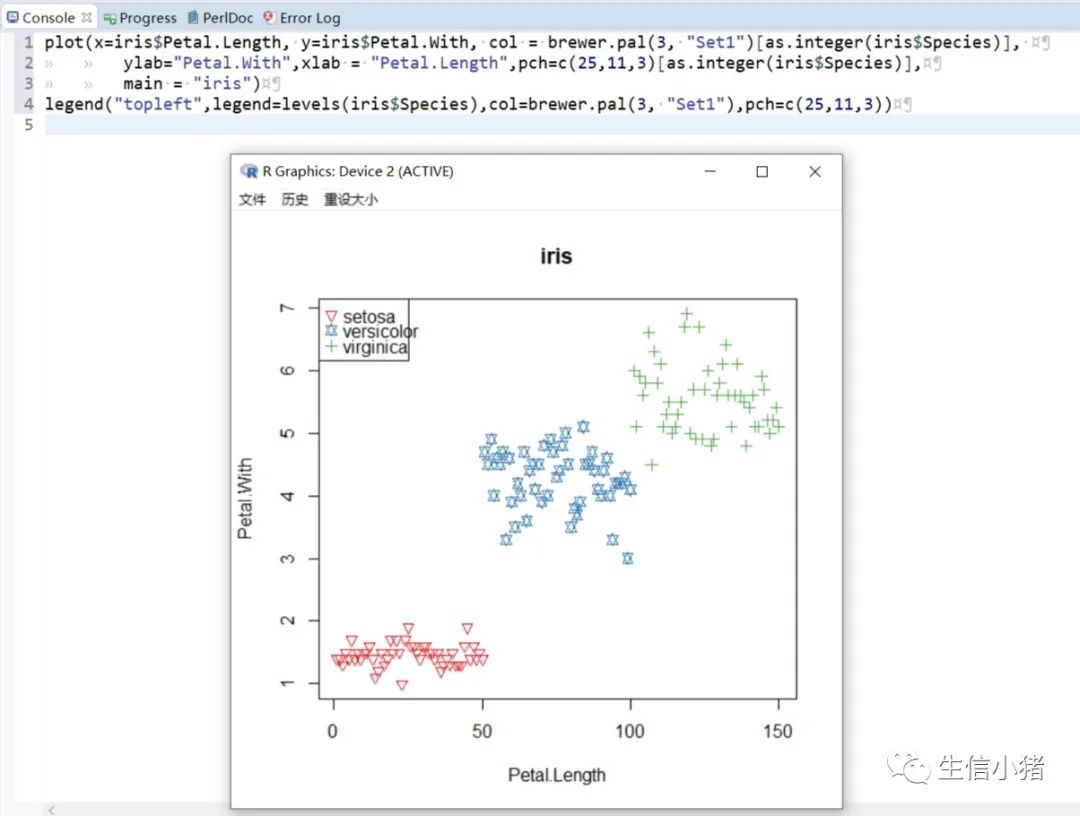
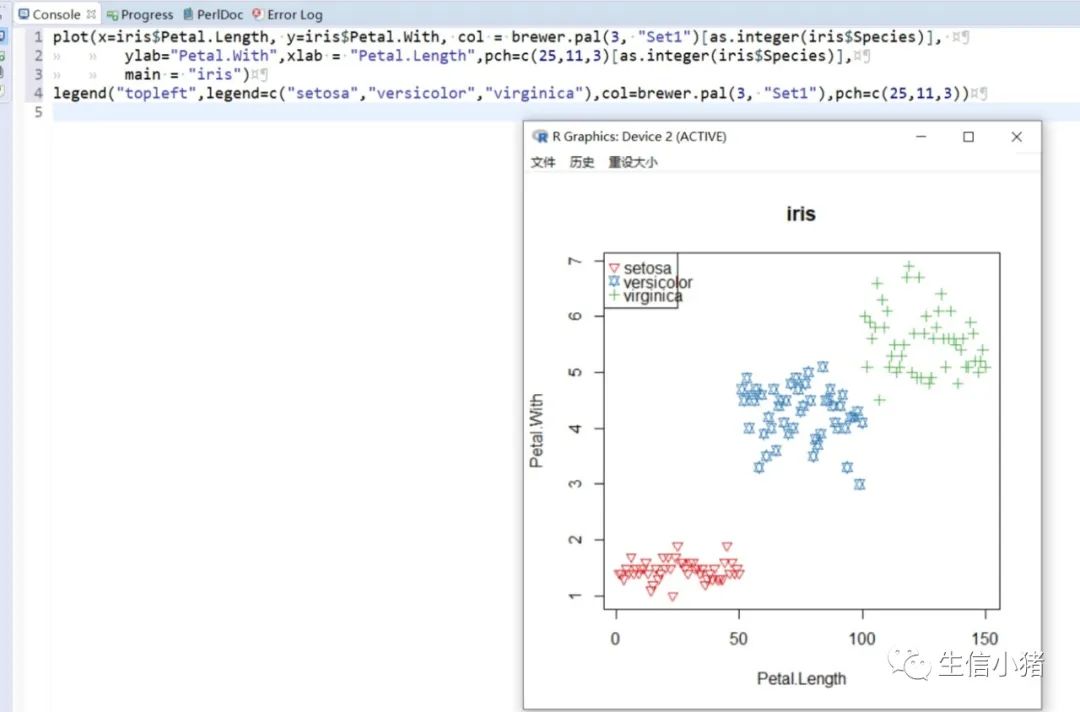
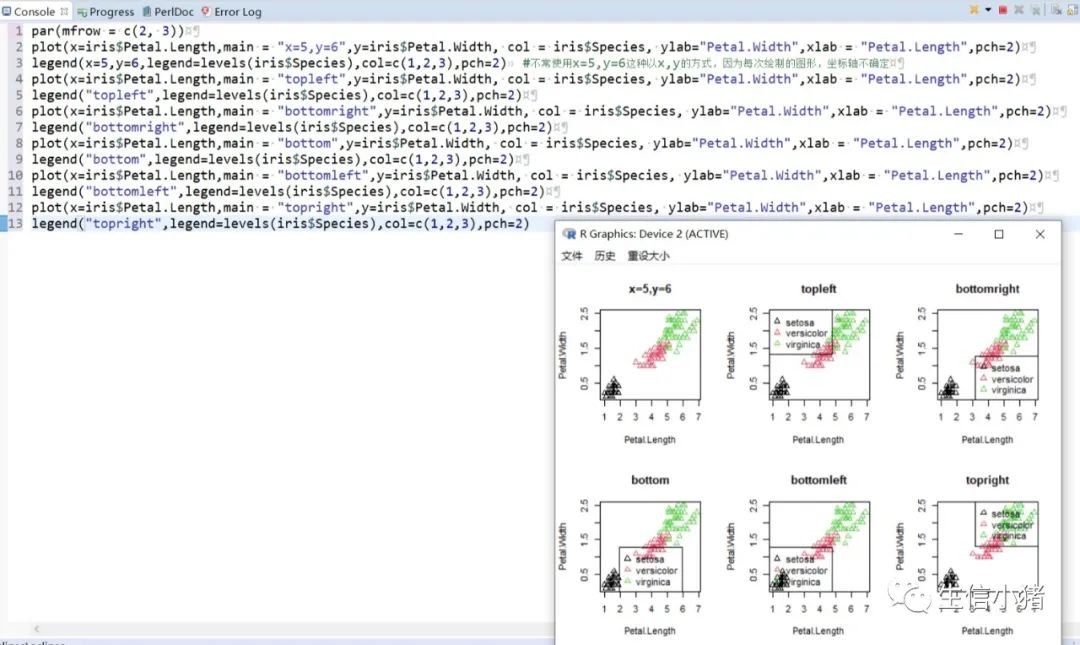
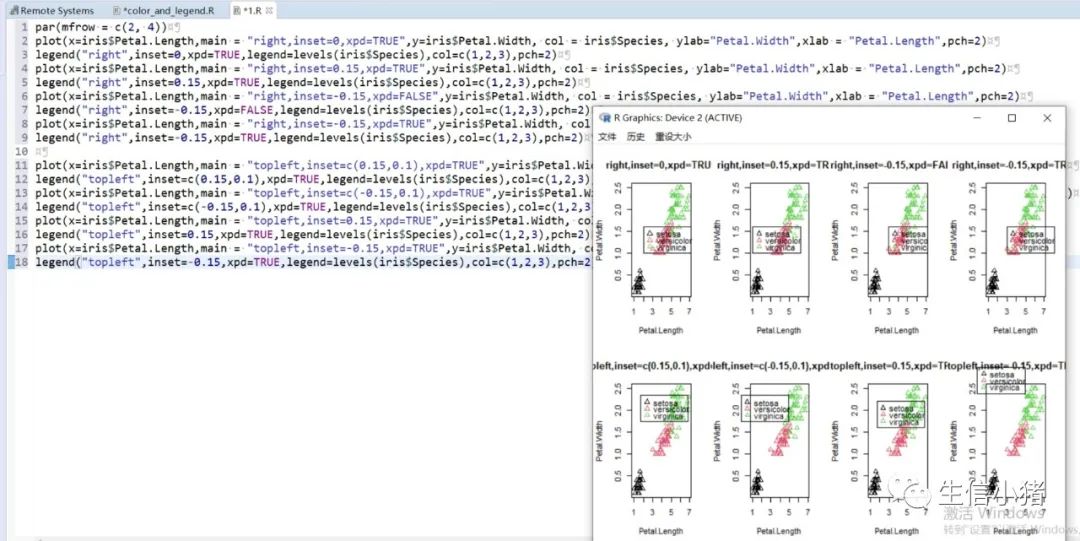
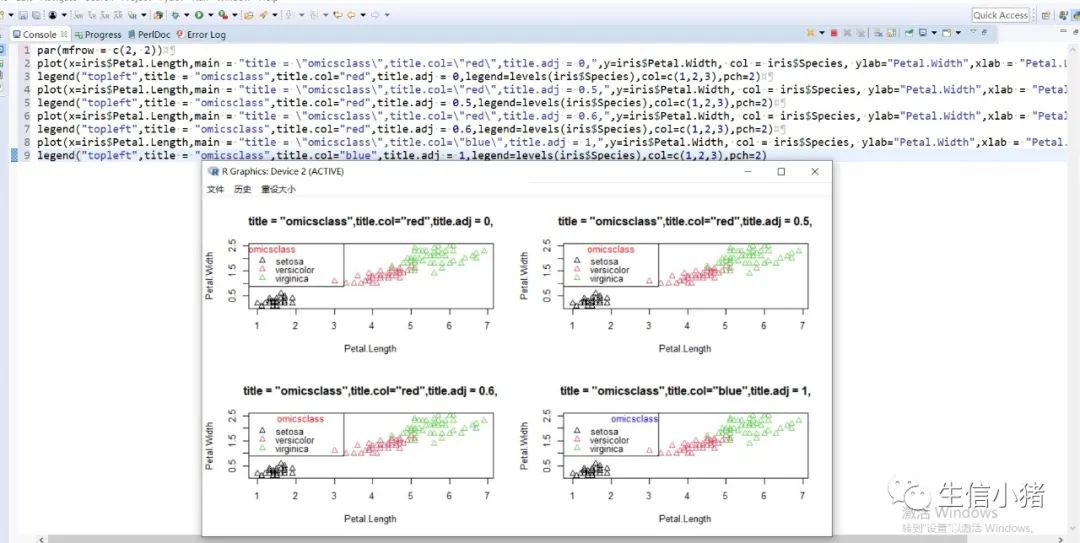
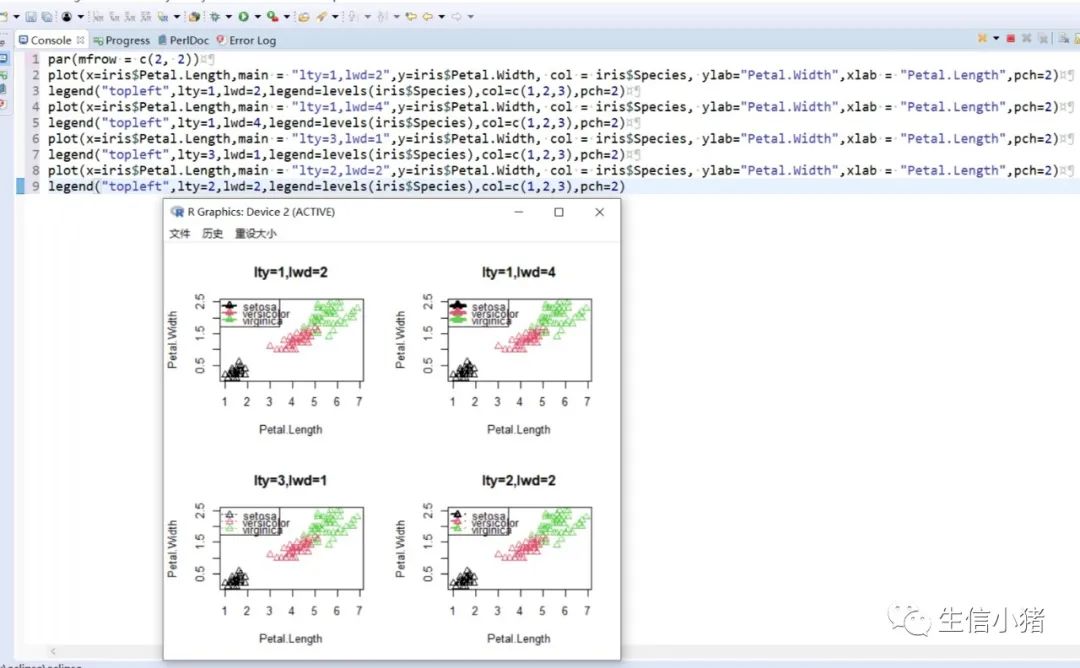
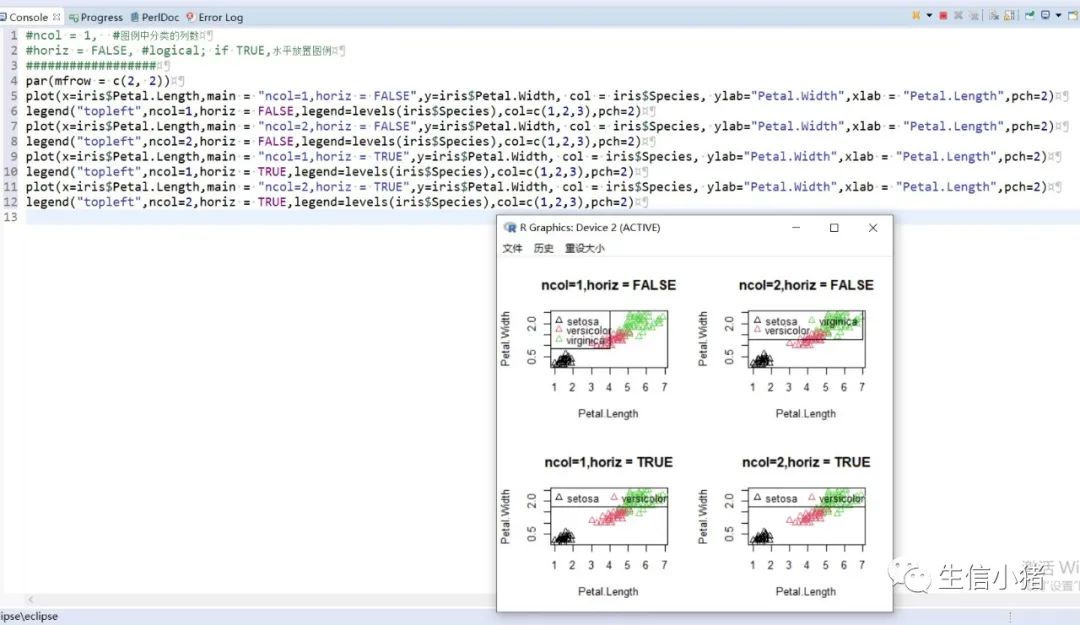
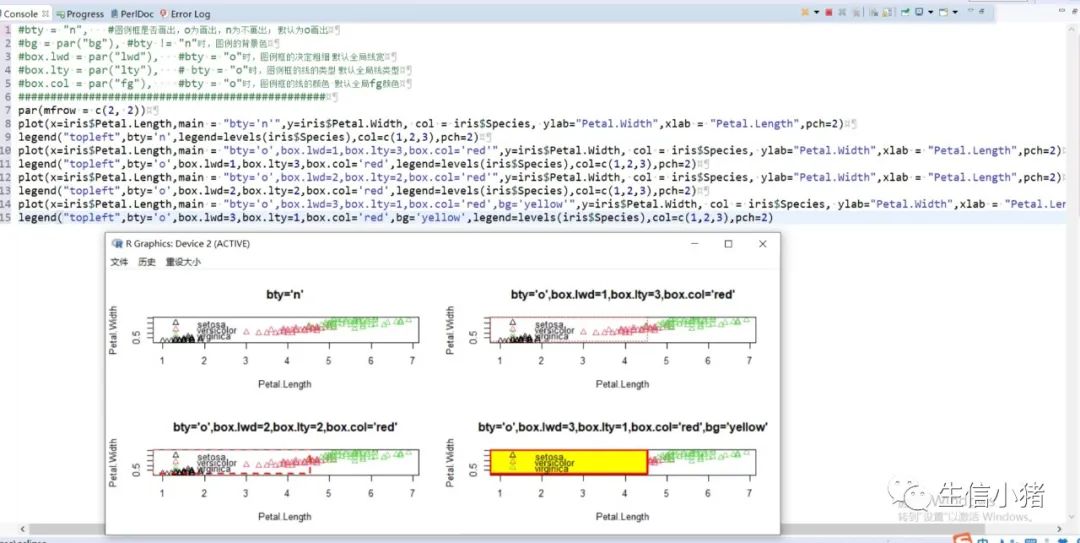
######################颜色应用 及添加legendplot(x=iris$Petal.Length, y=iris$Petal.With, col = brewer.pal(3, "Set1")[as.integer(iris$Species)],ylab="Petal.With",xlab = "Petal.Length",pch=c(25,11,3)[as.integer(iris$Species)],main = "iris") #产生颜色向量,标记每一个点的颜色#给图片添加图例legendlegend("topleft",legend=levels(iris$Species),col=brewer.pal(3, "Set1"),pch=c(25,11,3)) #这里面的levels(iris$Species)可以替换成c("setosa","versicolor","virginica")也可以达到同样的效果#图例用法及说明############################legend(x, y = NULL, #X,y用于定位图例,一般用单键词"bottomright", "bottom", "bottomleft", "left", "topleft", "top", "topright", "right" and "center" 代替x,ylegend, #字符向量col = par("col"), #图例中出现的点或线的颜色pch, #点的类型cex = 1, #字符 及点的大小lty, #图例中线的类型lwd, #图例中线的宽度ncol = 1, #图例中分类的列数horiz = FALSE, #logical; if TRUE,水平放置图例inset = 0, #0-1小数,相对边界偏移,正数远离边界,负数接近边界xpd=FALSE, #xpd=FALSE,即不允许在作图区域外作图。 # 剪切超出绘图区域部分;如果 xpd 值为 FALSE, 所有的图形被剪切为绘图区域 (plot); 如果 xpd 值为 TRUE, 所有图形被剪切为图像区域 (figure) (绘图区域和边缘 (margin)); 如果 xpd 值为NA, 则图形被剪切为设备区域, 即图像区域和外边界(outer) .adj = c(0, 0.5), #图例中字体的对齐相对位置,第一个数字水平相对位置,0表示水平左对齐,第二个为垂直相对位置对齐方式text.width = NULL, #图例字体所占的宽度text.col = par("col"), # 图例字体的颜色title = NULL, #给图例加标题title.col= "black", #标题颜色title.adj = 0.5, #图例标题的对齐相对位置,0.5为默认居中对齐。0左对齐,1为右对齐。bty = "n", #图例框是否画出,o为画出,n为不画出; 默认为o画出bg = par("bg"), #bty != "n"时,图例的背景色box.lwd = par("lwd"), #bty = "o"时,图例框的决定粗细 默认全局线宽box.lty = par("lty"), # bty = "o"时,图例框的线的类型 默认全局线类型box.col = par("fg"), #bty = "o"时,图例框的线的颜色 默认全局fg颜色fill = NULL, #用固定的颜色进行填充,通常绘制柱状图时使用,通常等于颜色向量border = "black", #当fill = 参数存在的情况下,填充色的边框spt.bg = NA, #点的背景色pt.cex = cex, #点的大小pt.lwd = lwd #点的边缘的线宽)par(mfrow = c(2, 3))plot(x=iris$Petal.Length,main = "x=5,y=6",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend(x=5,y=6,legend=levels(iris$Species),col=c(1,2,3),pch=2) #不常使用x=5,y=6这种以x,y的方式,因为每次绘制的图形,坐标轴不确定plot(x=iris$Petal.Length,main = "topleft",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "bottomright",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("bottomright",legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "bottom",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("bottom",legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "bottomleft",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("bottomleft",legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "topright",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topright",legend=levels(iris$Species),col=c(1,2,3),pch=2)#调整图例位置#inset = 0, #0-1小数,相对边界偏移,正数远离边界,负数接近边界#xpd=FALSE, #xpd=FALSE,即不允许在作图区域外作图。 # 剪切超出绘图区域部分;如果 xpd 值为 FALSE, 所有的图形被剪切为绘图区域 (plot); 如果 xpd 值为 TRUE, 所有图形被剪切为图像区域 (figure) (绘图区域和边缘 (margin)); 如果 xpd 值为NA, 则图形被剪切为设备区域, 即图像区域和外边界(outer) .par(mfrow = c(2, 4))plot(x=iris$Petal.Length,main = "right,inset=0,xpd=TRUE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("right",inset=0,xpd=TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "right,inset=0.15,xpd=TRUE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("right",inset=0.15,xpd=TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "right,inset=-0.15,xpd=FALSE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("right",inset=-0.15,xpd=FALSE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "right,inset=-0.15,xpd=TRUE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("right",inset=-0.15,xpd=TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "topleft,inset=c(0.15,0.1),xpd=TRUE",y=iris$Petal.With, col = iris$Species, ylab="Petal.With",xlab = "Petal.Length",pch=2)legend("topleft",inset=c(0.15,0.1),xpd=TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2) #inset=c(0.15,0.1),0.5表示水平方向的偏移,0.1表示垂直方向的偏移,都是正值表示都远离边界plot(x=iris$Petal.Length,main = "topleft,inset=c(-0.15,0.1),xpd=TRUE",y=iris$Petal.With, col = iris$Species, ylab="Petal.With",xlab = "Petal.Length",pch=2)legend("topleft",inset=c(-0.15,0.1),xpd=TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "topleft,inset=0.15,xpd=TRUE",y=iris$Petal.With, col = iris$Species, ylab="Petal.With",xlab = "Petal.Length",pch=2)legend("topleft",inset=0.15,xpd=TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "topleft,inset=-0.15,xpd=TRUE",y=iris$Petal.With, col = iris$Species, ylab="Petal.With",xlab = "Petal.Length",pch=2)legend("topleft",inset=-0.15,xpd=TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)#图例标题#title = NULL, #给图例加标题#title.col= "black", #标题颜色#title.adj = 0.5, #图例标题的对齐相对位置,0.5为默认居中对齐。0左对齐,1为右对齐。par(mfrow = c(2, 2))plot(x=iris$Petal.Length,main = "title = "omicsclass",title.col="red",title.adj = 0,",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",title = "omicsclass",title.col="red",title.adj = 0,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "title = "omicsclass",title.col="red",title.adj = 0.5,",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",title = "omicsclass",title.col="red",title.adj = 0.5,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "title = "omicsclass",title.col="red",title.adj = 0.6,",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",title = "omicsclass",title.col="red",title.adj = 0.6,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "title = "omicsclass",title.col="blue",title.adj = 1,",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",title = "omicsclass",title.col="blue",title.adj = 1,legend=levels(iris$Species),col=c(1,2,3),pch=2)#lty, #图例中线的类型#lwd, #图例中线的宽度par(mfrow = c(2, 2))plot(x=iris$Petal.Length,main = "lty=1,lwd=2",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",lty=1,lwd=2,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "lty=1,lwd=4",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",lty=1,lwd=4,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "lty=3,lwd=1",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",lty=3,lwd=1,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "lty=2,lwd=2",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",lty=2,lwd=2,legend=levels(iris$Species),col=c(1,2,3),pch=2)#ncol = 1, #图例中分类的列数#horiz = FALSE, #logical; if TRUE,水平放置图例##################par(mfrow = c(2, 2))plot(x=iris$Petal.Length,main = "ncol=1,horiz = FALSE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",ncol=1,horiz = FALSE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "ncol=2,horiz = FALSE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",ncol=2,horiz = FALSE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "ncol=1,horiz = TRUE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",ncol=1,horiz = TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "ncol=2,horiz = TRUE",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",ncol=2,horiz = TRUE,legend=levels(iris$Species),col=c(1,2,3),pch=2)#bty = "n", #图例框是否画出,o为画出,n为不画出; 默认为o画出#bg = par("bg"), #bty != "n"时,图例的背景色#box.lwd = par("lwd"), #bty = "o"时,图例框的决定粗细 默认全局线宽#box.lty = par("lty"), # bty = "o"时,图例框的线的类型 默认全局线类型#box.col = par("fg"), #bty = "o"时,图例框的线的颜色 默认全局fg颜色################################################par(mfrow = c(2, 2))plot(x=iris$Petal.Length,main = "bty='n'",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",bty='n',legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "bty='o',box.lwd=1,box.lty=3,box.col='red'",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",bty='o',box.lwd=1,box.lty=3,box.col='red',legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "bty='o',box.lwd=2,box.lty=2,box.col='red'",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",bty='o',box.lwd=2,box.lty=2,box.col='red',legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "bty='o',box.lwd=3,box.lty=1,box.col='red',bg='yellow'",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",bty='o',box.lwd=3,box.lty=1,box.col='red',bg='yellow',legend=levels(iris$Species),col=c(1,2,3),pch=2)#pt.bg = NA, #点的背景色#pt.cex = cex, #点的大小#pt.lwd = lwd #点的边缘的线宽###############################par(mfrow = c(2, 2))plot(x=iris$Petal.Length,main = "pt.bg=NA,pt.cex=1,pt.lwd=1,pch=2",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",pt.bg=NA,pt.cex=1,pt.lwd=1,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "pt.bg='red',pt.cex=1,pt.lwd=2,pch=2",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=2)legend("topleft",pt.bg='yellow',pt.cex=1,pt.lwd=2,legend=levels(iris$Species),col=c(1,2,3),pch=2)plot(x=iris$Petal.Length,main = "pt.bg=NA,pt.cex=2,pt.lwd=1,pch=21",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=21)legend("topleft",pt.bg=NA,pt.cex=2,pt.lwd=1,legend=levels(iris$Species),col=c(1,2,3),pch=21)plot(x=iris$Petal.Length,main = "pt.bg='yellow',pt.cex=2,pt.lwd=2,pch=21",y=iris$Petal.Width, col = iris$Species, ylab="Petal.Width",xlab = "Petal.Length",pch=21)legend("topleft",pt.bg='yellow',pt.cex=2,pt.lwd=2,legend=levels(iris$Species),col=c(1,2,3),pch=21)##################练习:绘制点线图,并添加legend###dev.off()x<-1:10y1=x*xy2=2*y1par(mar(5,4,4,5)+0.1)plot(x, y1, type="b", pch=11, col="red", xlab="x", ylab="y",main="my legend set")# Add a linelines(x, y2, pch=18, col="blue", type="b", lty=2)# Add a legendlegend("top", legend=c("Line 1", "Line 2"),inset = -0.05,xpd=TRUE,horiz = T,bty='n',col=c("red", "blue"), lty=1:2, cex=0.8,pch=c(11,18))
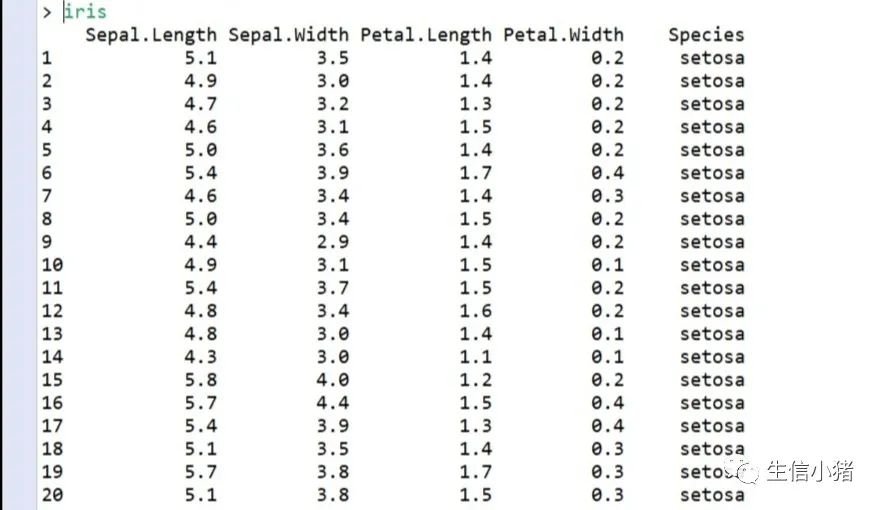
其中iris这个内置的数据的第Species列有三个不同的因子,分别为setosa,versicolor,virginica,这里由于篇幅有限,所以就截取了前20行,以至于不能看到第Species列的其他因子

以上是关于R语言颜色-图例legend及位置调整的主要内容,如果未能解决你的问题,请参考以下文章