R语言数据分析之ClusterProfiler
Posted 蛋白组学数据分析
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了R语言数据分析之ClusterProfiler相关的知识,希望对你有一定的参考价值。
library(xlsx)library(ggplot2)library(stringr)library(clusterProfiler)library(org.Hs.eg.db)#根据分析的物种选择setwd("D:/Rfiles/RLanguage/Clusterprofiler")gene <- read.table("gene.txt",header=T,sep=" ")#ID转换ID <- bitr(gene$Name, fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")#OrgDb修改成相应的分析物种,后续类似#GO分开进行富集BP <- enrichGO(ID$ENTREZID, "org.Hs.eg.db", keyType = "ENTREZID",ont = "BP",pvalueCutoff = 0.05,pAdjustMethod = "BH",qvalueCutoff = 0.1, readable=T)MF <- enrichGO(ID$ENTREZID, "org.Hs.eg.db", keyType = "ENTREZID",ont = "MF",pvalueCutoff = 0.05,pAdjustMethod = "BH",qvalueCutoff = 0.1, readable=T)CC <- enrichGO(ID$ENTREZID, "org.Hs.eg.db", keyType = "ENTREZID",ont = "CC",pvalueCutoff = 0.05,pAdjustMethod = "BH",qvalueCutoff = 0.1, readable=T)#pathway分析ekk <- enrichKEGG(ID$ENTREZID, keyType = "kegg", organism = "hsa", pAdjustMethod = "BH", pvalueCutoff = 0.05, qvalueCutoff = 0.05, minGSSize = 2)#geneID转gene Symboly <- setReadable(ekk, 'org.Hs.eg.db', keyType = "ENTREZID")#ekk根据情况改成BP、MF、CC,主要是看想要导出哪个结果c(nrow(y))write.xlsx(y, sheetName = "KEGG", file = "result.xlsx")#这里的KEGG根据情况改成BP、MF、CC#柱形图与气泡图展示barplot(KEGG, showCategory = 15, title = "Enrichment Pathway")#title可修改成BP、MF、CC、Pathway# Save as PDF, 12*10 inchesdotplot(KEGG, showCategory = 15, title = "Enrichment Pathway")#title可修改成BP、MF、CC、Pathway# Save as PDF, 9*8 inches
-
数据准备:
图1 数据准备
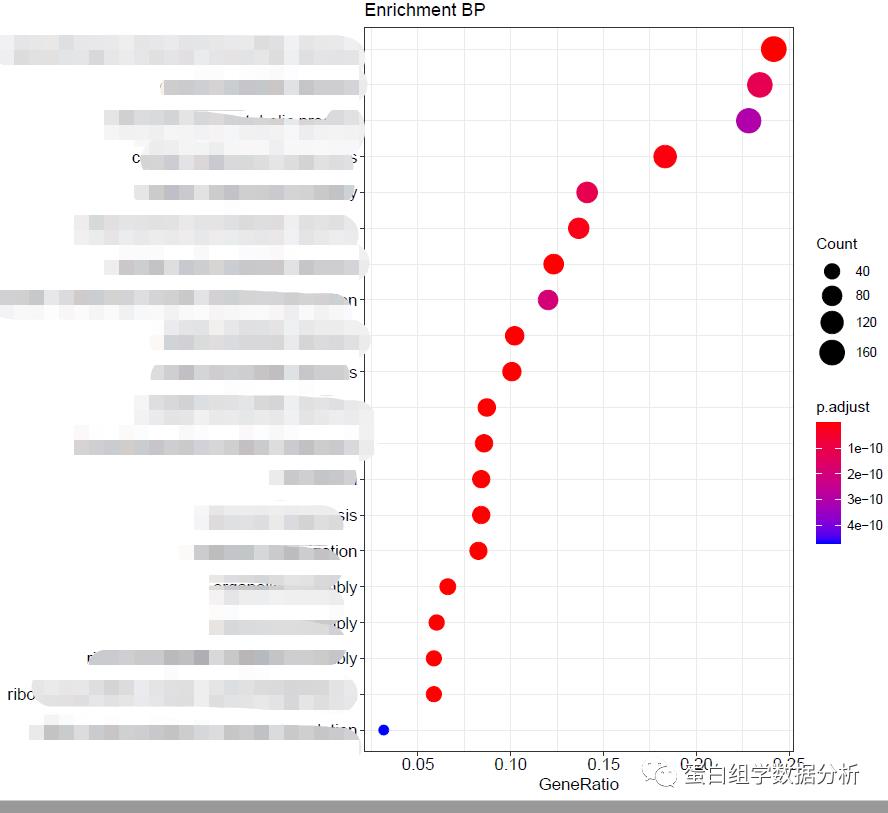
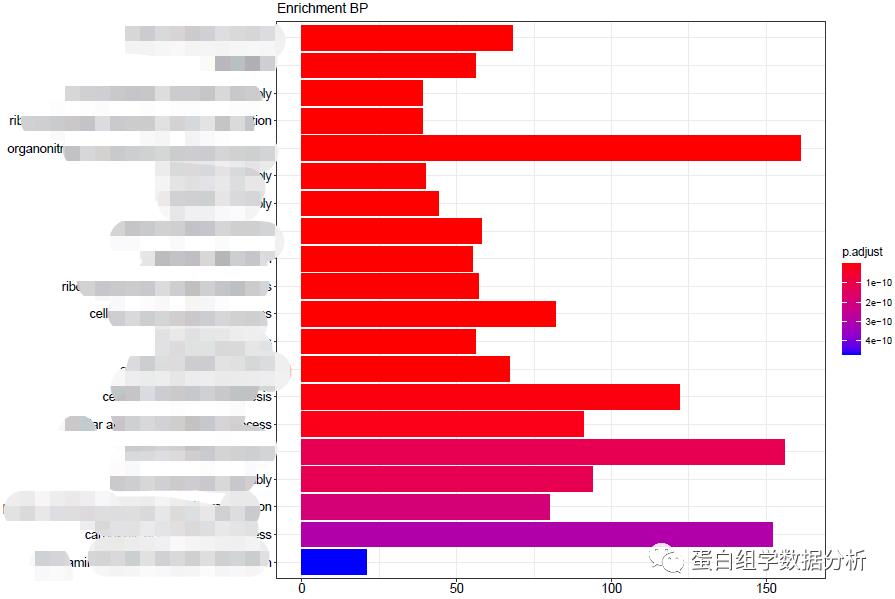
-
往期生信分析导航:
参考资料:
南方医科大学余光创教授
http://portal.smu.edu.cn/jcyxy/info/1084/2203.htm
ClusterProfiler,该R包版本在不断更新,实现了GO和KEGG等分析,同时可进行数据可视化。
http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html
Bioconductor上提供了以下19个物种的Org类型的包,包含了这些物种的GO注释信息
https://cloud.tencent.com/developer/article/1625246
以上是关于R语言数据分析之ClusterProfiler的主要内容,如果未能解决你的问题,请参考以下文章