瞧!你除了Excel,还有Perl可以用
Posted 牛学汇
tags:
篇首语:本文由小常识网(cha138.com)小编为大家整理,主要介绍了瞧!你除了Excel,还有Perl可以用相关的知识,希望对你有一定的参考价值。
再不点蓝字关注,机会就要飞走了哦
俗话说的好
“世界上没有一个perl程序不能解决的事情,如果有,那就两个或N个。”
呐!慈禧太后不小心弄断指甲的那些日子,都是靠金牛角度过的
小编最近处理数据时也是被Excel折了翅膀,但小编是靠perl度过的

来:直接上代码
#!/usr/bin/perl
# Searcher for aminoacid patterns
# Ask the user the patterns for search
print "Please, introduce the pattern to search in query.seq: ";
$patron = <STDIN>;
chomp $patron;
# Open the database file
# but if it can't it ends the program
open (query, "protein.fasta") || die "problem opening the file query_seq.txt ";
# Look line by line the SWISS-PROT sequence
while (<query>) {
chomp $_;
# When arrives to the SQ field,put the mark in 1
if ($_ =~ /^SQ/) {
$signal_seq = 1;
# When arrive to the end of sequence, leave the curl
# Check that this expression is put before to check
# the mark=1,because this line doesn't belong to the aminoacid sequence
} elsif ($_ =~ /^///) {
last;
# Check the mark if it is equal to 1, if possitive
# eliminate the blank spaces in the sequence line
# and join every line in a new variable
# To concatenate, we also can do:
# $secuencia_total.=$_;
} elsif ($signal_seq == 1) {
$_ =~ s/ //g;
$secuencia_total=$secuencia_total.$_;
}
}
# Now check the sequence, collected in its entirety,
# for the given pattern
if ($secuencia_total =~ /$patron/) {
print "The sequence query.seq contains the pattern $patron
";
} else {
print "The sequence query.seq doesn't contains the pattern $patron
";
}
# Finally we close the file
# and leave the program
close (query);
exit;
![]() 此代码用来查找氨基酸模式
此代码用来查找氨基酸模式
文件格式要这样准备
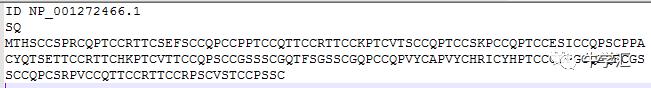
我只是个分割线
![]()
perl还可以根据ID号提取蛋白质或氨基酸序列、计算氨基酸频率,批量提取genebank序列文件中编码区、CDS长度、氨基酸等信息.....包括你想要的数据挖掘,文本处理。
以上是关于瞧!你除了Excel,还有Perl可以用的主要内容,如果未能解决你的问题,请参考以下文章